Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Splice-site variants in X-linked IKBKG cause NEMO-deleted exon5 autoinflammatory syndrome (NEMO-NDAS); a pseudogene (IKBKGP1) complicates genetic diagnosis. NEMO-NDAS is four times more common in girls than boys and patients (pts) experience systemic inflammation, panniculitis, non-cirrhotic portal hypertension and splenomegaly, that are only partially responsive to TNF and/or Jak inhibition. Childhood mortality is high (~19%). We aimed to characterize the complex genetics in NEMO-NDAS.
Methods: Pts were enrolled in natural history protocol NCT02974595. Specific primers were used for targeted long-range PCR of IKBKG and IKBKGP1 in pts and family members. Variants were validated by Sanger or amplicon deep sequencing. Serum cytokines were measured. Flow cytometry (FC) was used to analyze and sort T cell subsets and variant allele fraction (VAF), exon 5 skipping and X-inactivation were assessed in these cells. RNA-seq of whole blood (WB), sorted cells, liver, and skin was conducted to validate the genetic model.
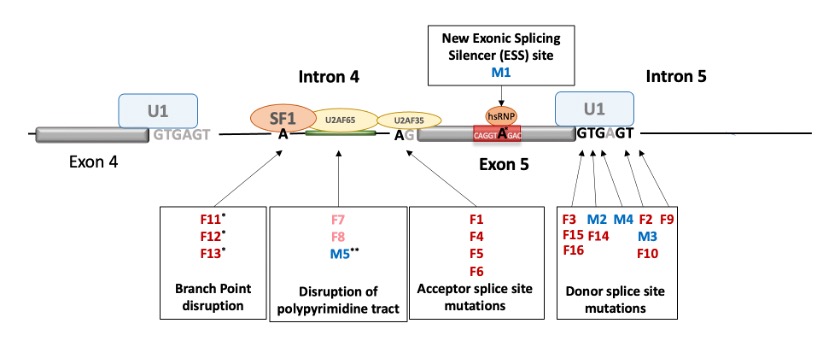
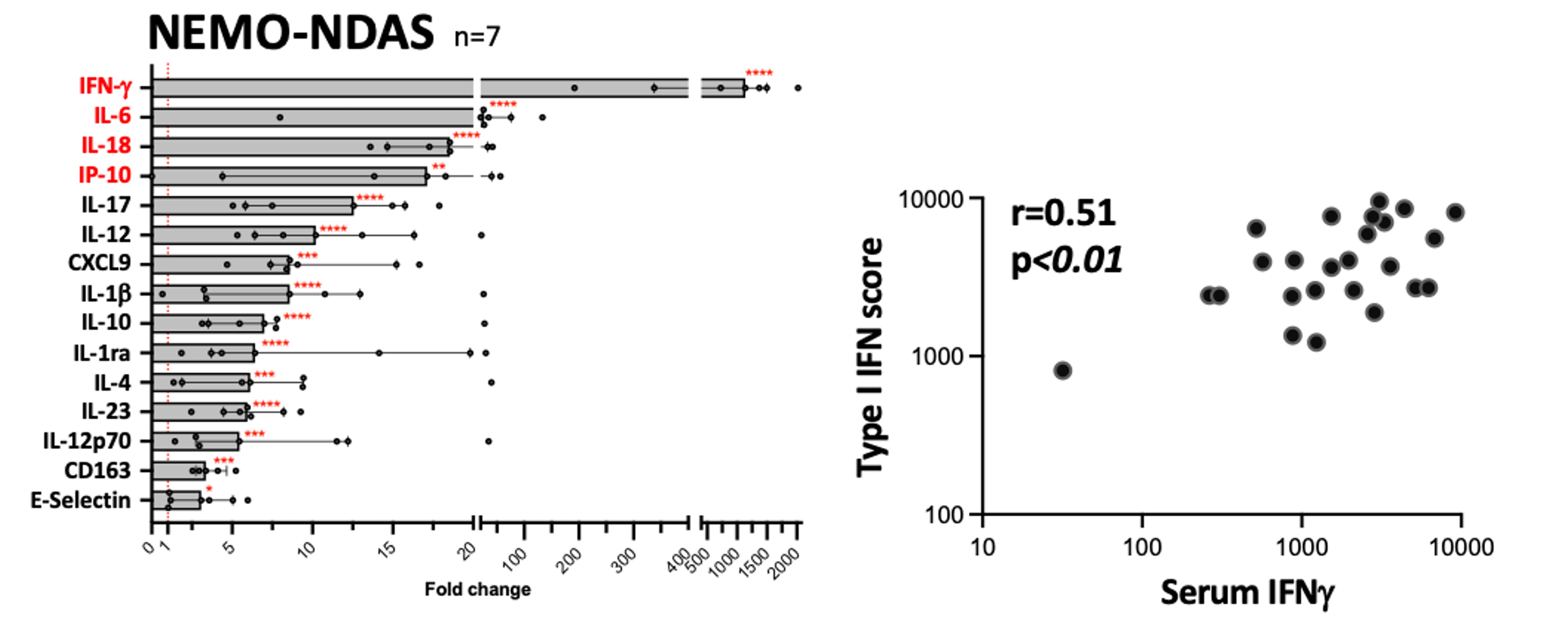
Results: Twenty-one pts (16 females, 13 sporadic, 3 familial, and 5 males) had 12 different splice-site variants in IKBKG (Fig 1). Of the boys, 1 pt (M5) with Klinefelter syndrome (XXY karyotype) had a polypyrimidine tract mutation that changed the branch point, 1 had a germline exonic mutation leading to a leaky splice-site and 3 had mosaic canonical splice-site mutations. Of the girls, 11 pts with canonical splice-site mutations and the 3 familial pts (a mother and 2 daughters) with a branch point mutation were germline, while 2 mosaic pts had a polypyrimidine tract deletion (F7) or insertion (F8). In all girls with germline de novo mutations tested, the variant was on the paternal X chromosome. Pt F14 inherited a splice-site variant in IKBKGP1 from her father, which was likely repositioned to IKBKG, highlighting non-allelic homologous recombination (NAHR) between the gene and pseudogene. Screening of the 1000 Genomes database found 6 subjects with a splice-site variant in IKBKGP1 exon 5 region. To identify the cellular source of the high serum IFNγ levels observed (Fig 2), a customized FC panel (n=6) was run and showed significant expansion of activated TCRγδ+ T (γδT) cells that expressed mostly wildtype IKBKG message with exon 5 skipping ranging from 0 to 7.6% in comparison to 15.3 to 74.6% in WB. In mosaic pts, the activated cells were wildtype. In activated γδT cells from girls with germline mutations, SNP analysis suggested skewed ( >65%) X-inactivation of the mutant allele suggesting selection pressure favoring wildtype message. Activated γδT cells produced IFNγ upon PMA stimulation and were increased in inflamed skin and liver tissues.
Conclusion: Our data suggest that NEMO-NDAS-causing variants in canonical splice-sites can be present as germline mutations with X-inactivation in girls or present as mosaic mutations in males. The pseudogene can harbor splice-site mutations that can undergo NAHR at the IKBKG/IKBKGP1 locus and increase the chance to inherit the mutation from the paternal germline. The expansion of γδT cells and their skewed X-inactivation raise questions on their role in the pathogenesis of NEMO-NDAS and may provide a therapeutic target for better treatment.
*, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001.
To cite this abstract in AMA style:
Almeida de Jesus A, Friend K, Lin B, Karlins E, McNinch C, Alehashemi S, Kauffman K, BHUYAN F, Newbolt T, Bohrer A, Rastegar A, Park S, Kahle D, Mitchell J, Chen A, Torreggiani S, Cetin Gedik K, Uss K, Khojah A, Wu E, Scott C, Leahy T, MacDermott E, Killeen O, Arkachaisri T, Nolan B, Gucev Z, Cook K, Mammadova V, Nasrullayeva G, Correia Marques M, Bosk A, Ozen S, Canna S, Tusseau M, Chopin E, Boursier G, Hentgen V, Elhani I, Sestan M, Jelusic M, Fink D, Kuhns D, Dalgard C, Belot A, Moran T, Meyer-Barber K, Oler A, Barber D, Goldbach-Mansky R. Divergent Genetic Architecture in Boys and Girls with NEMO-deleted Exon 5 Autoinflammatory Syndrome (NEMO-NDAS) Implies Role for Wildtype Effector Cells [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/divergent-genetic-architecture-in-boys-and-girls-with-nemo-deleted-exon-5-autoinflammatory-syndrome-nemo-ndas-implies-role-for-wildtype-effector-cells/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/divergent-genetic-architecture-in-boys-and-girls-with-nemo-deleted-exon-5-autoinflammatory-syndrome-nemo-ndas-implies-role-for-wildtype-effector-cells/