Session Information
Session Type: ACR Abstract Session
Session Time: 2:30PM-4:00PM
Background/Purpose: Our understanding of disease pathogenesis in Rheumatoid Arthritis (RA) is rapidly expanding with the advent of ‘Omics’ techniques. Importantly, current analysis takes into account many clinical covariates such as gender and age, however there are no studies to date that have systematically examined the impact that the timing of sample acquisition has on transcriptional profiles. This is a fundamental covariate given the real-world clinical setting of samples acquisition. Moreover, within day variation in RA symptoms are characteristic of disease (e.g. early morning stiffness). Herein, we evaluated the diurnal variation in transcriptomic profiles in 10 RA patients.
Methods: Whole blood was drawn into Paxgene tubes from 10 RA patients over the course of one day (8am, 9am, 11am, 1pm and 3pm), and stored at -80°C. RNA was extracted using the Paxgene Blood miRNA kit and libraries prepared for RNA-Seq using the Illumina TruSeq Stranded Total RNA Ribo-Zero H/M/R Gold kit. RNA-seq was performed and data analysed using the likelihood ratio test in the DESeq2 R package to identify any genes that showed a change in expression across the different time points. The genes identified as being differentially expressed and having a fold change greater than 1.5 between any two time points in the RNA-Seq analysis were clustered to identify patterns of expression. Gene ontology enrichment analysis as well as pathway over-representation analysis using the Reactome database was performed.
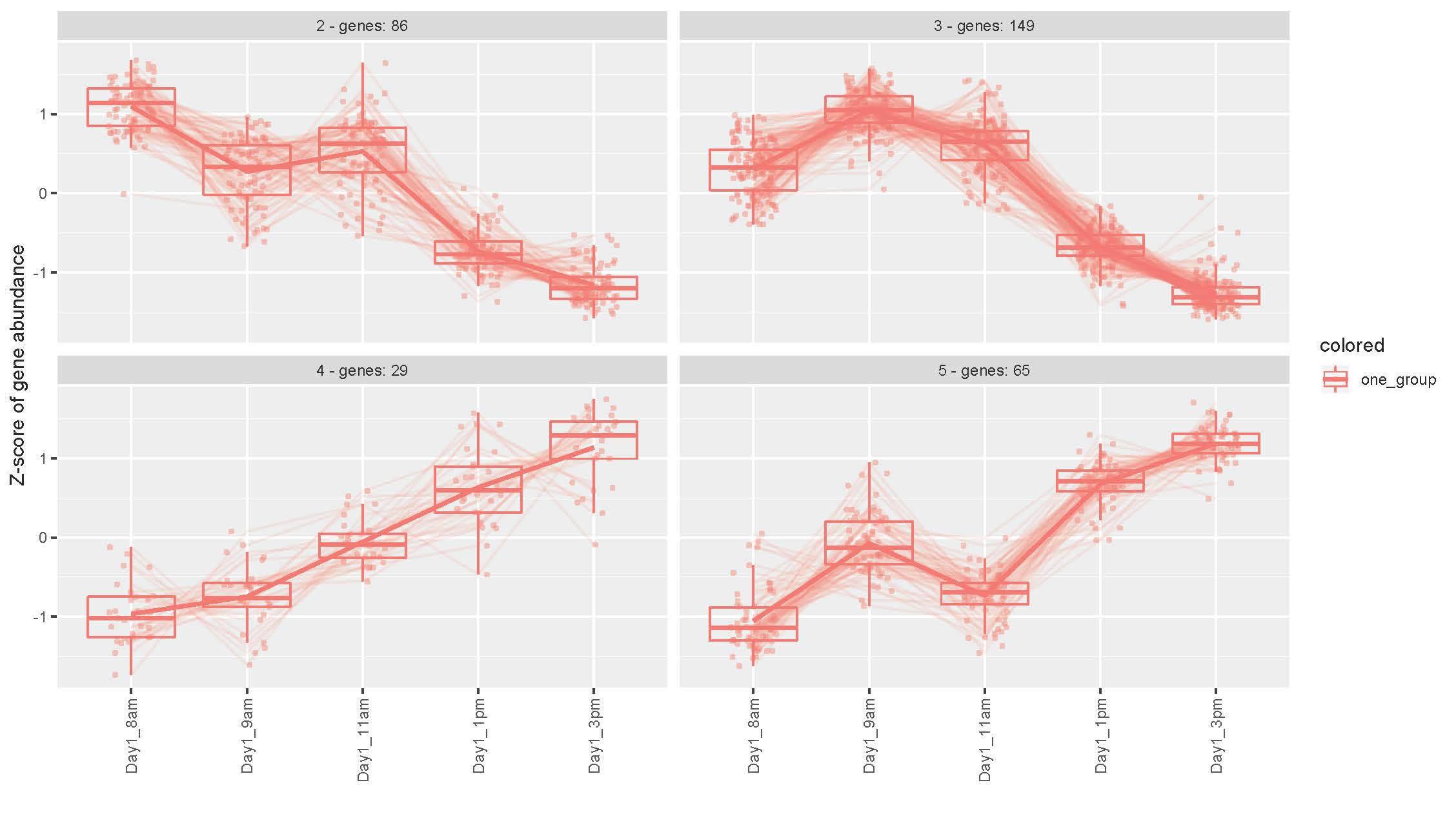
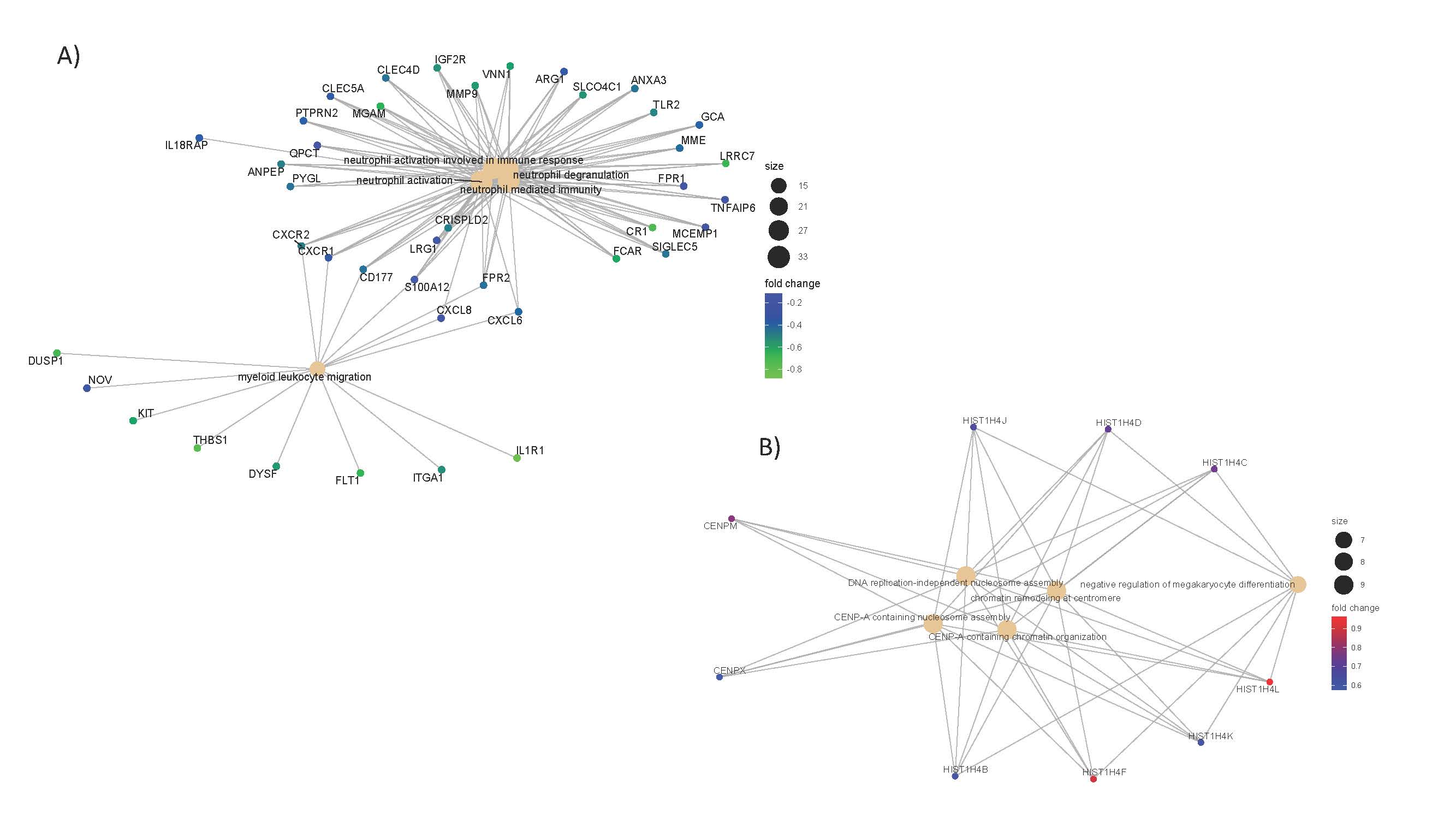
Results: RNA-Seq analysis identified 395 genes with a fold change of 1.5 or greater that were differentially expressed between the timepoints. Cluster analysis of these genes identified four patterns of expression. Two of the expression clusters (utilising 86 or 149 genes respectively) showed a decrease in transcripts throughout the day, figure 1. Notably, the gene ontology terms associated with the clusters were; neutrophil activation, myeloid and leucocyte migration, neutrophil degranulation, figure 2a. Furthermore, Reactome Pathways Analysis highlighted; interleukin-33 signalling, interleukin-10 signalling, and interleukin-1 family signalling as over-representated in the clusters. All of which are commensurate with increased immune activation in the morning. In comparison, the other two expression clusters (utilising 29 and 65 gene respectively) increased during the day, figure 1. These clusters were associated with DNA replication and processing pathways and gene ontology terms, figure 2b.
Conclusion: Our results clearly demonstrate that timing of sample collection has a significant impact on the transcription profile of circulating cells and should therefore be accommodated within biomarker studies. Moreover, alterations were observed across rather fundamental biological pathways that may inform disease pathogenesis studies in future.
To cite this abstract in AMA style:
Bennett L, Morton F, Fragoulis G, Paterson C, Rimmer D, Semple G, Young A, Nijjar J, Barrett M, Siebert S, Porter D, Goodyear C, McInnes I. Diurnal Stability of Transcriptional Profiles in Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/diurnal-stability-of-transcriptional-profiles-in-rheumatoid-arthritis/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/diurnal-stability-of-transcriptional-profiles-in-rheumatoid-arthritis/