Session Information
Title: Systemic Sclerosis, Fibrosing Syndromes and Raynaud's - Pathogenesis, Animal Models and Genetics
Session Type: Abstract Submissions (ACR)
Background/Purpose: To examine the heterogeneity of global transcriptome patterns in systemic sclerosis (SSc) skin from a large cohort of patients and controls.
Methods: Skin biopsies from 61 patients (70.5 % diffuse cutaneous involvement) enrolled in the GENISOS cohort or at the baseline visit of an imatinib study, as well as 36 unaffected controls of similar demographic background, were examined by Illumina HT-12 gene expression arrays. Follow-up q-PCR and immunohistochemistry experiments were also performed. Using a novel analytic approach based on expression profiles, we investigated how heterogeneity within SSc samples relates to specific disease-relevant cell types (e.g. fibroblasts or macrophages).
Results: We identified 2754 differentially expressed transcripts in SSc patients compared to controls. Clustering analysis revealed two prominent transcriptomes in SSc patients: Keratin and fibro-inflammatory signatures. Higher keratin transcript scores were associated with shorter disease duration and interstitial lung disease while higher fibro-inflammatory scores were associated with diffuse cutaneous involvement, higher skin score at biopsy site and higher modified Rodnan Skin Score. There were no significant associations with disease-related autoantibodies or concomitant treatment with immunosuppressive agents. A subgroup of patients with significantly longer disease duration had a normal-like transcript pattern.
Further analysis and immunohistochemistry staining indicated that the above-mentioned keratin signature was not a general marker of keratinocyte activation, but was in fact associated with an activation pattern in hair and adnexal structures.
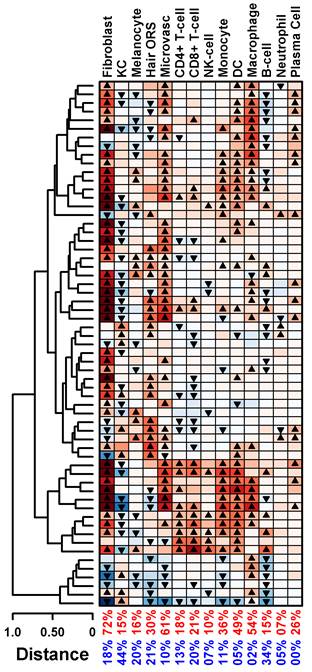
As shown in Figure 1, analysis of cell type-specific signature scores revealed remarkable heterogeneity across patients (each row represents a patient sample). Significantly high scores were observed in the majority of patients for fibroblasts (72% of patients), microvascular (61%), and macrophages (54%). The majority of samples with significant fibroblast scores (35 of 44 = 80%) also had significantly increased macrophage and/or dendritic cell scores. Only a minority of samples showed significantly high CD4+ T-cell, CD8+ T-cell and plasma cell scores (18%, 21%, and 26%).
Conclusion: In this large gene expression data set, a prominent keratin signature was present in addition to a fibro-inflammatory signature, supporting the notion that molecular dysregulations in SSc skin are not confined to the dermal layer but are in fact present in several skin compartments. Furthermore, the novel cell-specific transcript analysis showed significant heterogeneity of inflammatory profiles from SSc skin, which might be useful for stratifying patients for targeted treatment and/or predicting their response to immunosuppression.
Disclosure:
S. Assassi,
None;
W. Swindell,
None;
M. Wu,
None;
F. K. Tan,
None;
D. Khanna,
None;
D. E. Furst,
None;
D. Tashkin,
None;
R. Jahan-Tigh,
None;
M. D. Mayes,
None;
J. Gudjonsson,
None;
J. T. Chang,
None.
« Back to 2014 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/dissecting-the-heterogeneity-of-skin-gene-expression-patterns-in-systemic-sclerosis/