Session Information
Date: Monday, November 18, 2024
Title: SpA Including PsA – Diagnosis, Manifestations, & Outcomes Poster III
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Delays in diagnosis of psoriatic arthritis (PsA) are associated with indolent and non-specific signs and symptoms, and often result in physical impairment and poorer global health outcomes in the long term [1,2,3,4]. Digital tools such as machine learning algorithms have the potential to address diagnostic delays by identifying patients earlier in their disease course [5]. The purpose of this study was to develop and evaluate a novel machine learning model for identifying patients at risk of having undiagnosed PsA among patients who saw primary care services at Mayo Clinic.
Methods: Patients receiving primary care services (n=395,918 patients) at Mayo Clinic between 2012 and 2022 were split into training and testing sets. Cases with incident PsA (n=326) and controls with no evidence of PsA (n=395,592) were identified in both sets. For model training, each patient was assigned a prediction date, which preceded the initial diagnosis of PsA for cases, and was randomly assigned to controls. A gradient boosted trees algorithm was trained using electronic medical record (EMR) data documented during the two years prior to each patient’s prediction date. Input features included age, sex, diagnosis codes, medication prescriptions and laboratory results. The model was then evaluated in the test set on predictions made on 1 January 2018 with input data from the preceding two years. Area under the curve (AUC) was used to assess the model’s ability to discriminate between new cases of PsA diagnosed during and after 2018 and controls.
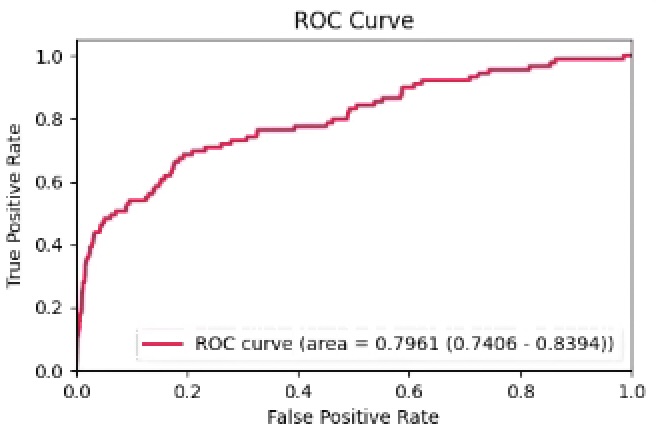
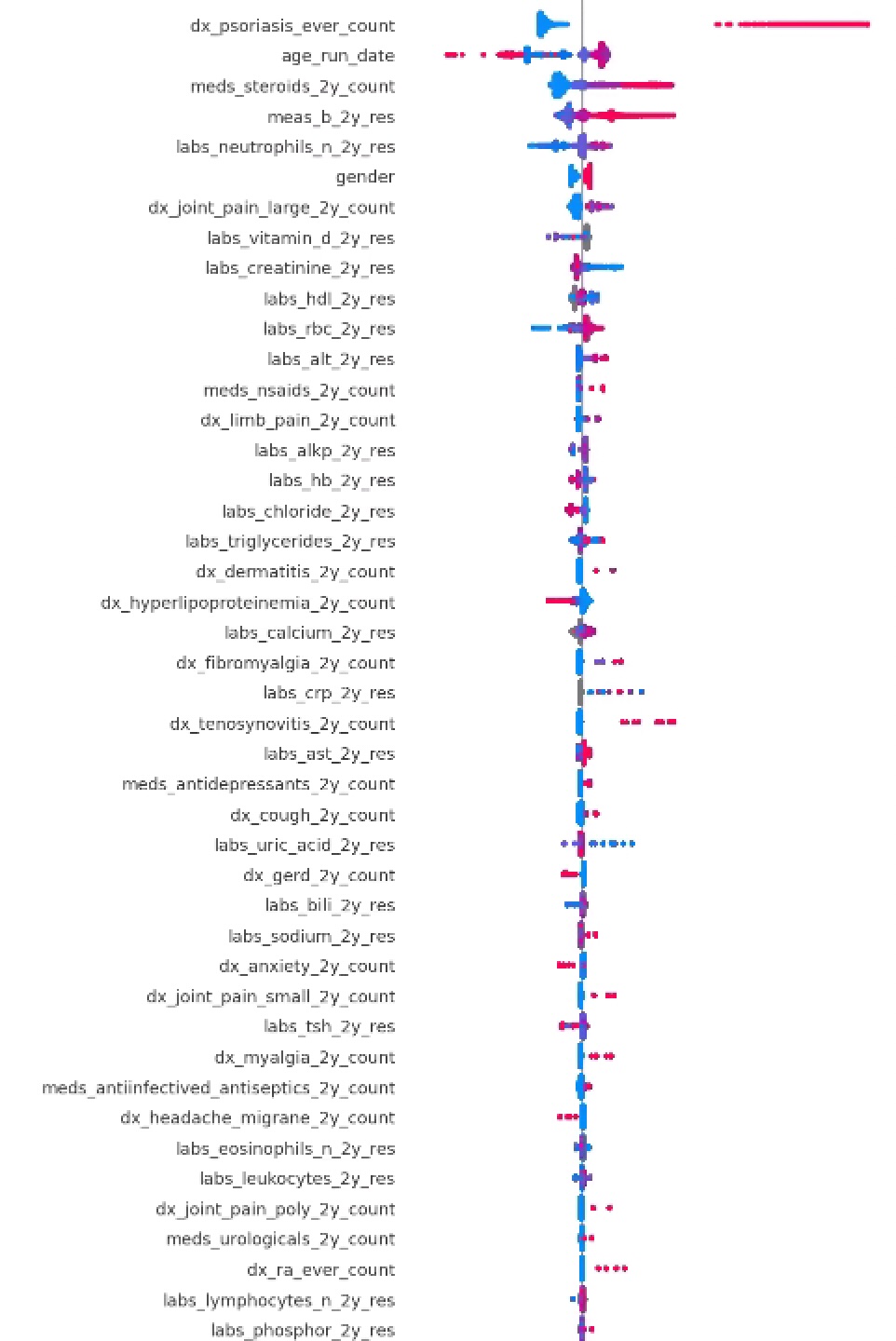
Results: The testing set included 89 patients with PsA (60 female; mean age 54.2, standard deviation [SD] 14.0) and 284,480 control patients (167,018 females; mean age 51.9, SD 18.2). The AUC on the test was 79.6% (fig 1). The top predictive features preceding the initial PsA diagnosis include diagnoses of psoriasis, joint pain and tenosynovitis, glucocorticoid and NSAID use, high body mass index, elevated CRP, ALT, triglycerides and decreased HDL (fig 2).
Conclusion: The model displayed good performance in its ability to discriminate between cases of PsA and controls. The model selected input features associated with preexisting psoriasis, joint inflammation and an adverse metabolic profile [6]. Implementation of the model may help identify patients with undiagnosed PsA in the primary care population using EMR. Improving time to diagnosis could help patients receive treatment and reduce downstream sequelae from untreated disease.
To cite this abstract in AMA style:
Dreyfuss M, Riesel D, Shalev G, Getz B, Ramni O, Jenudi Y, Duchovny D, Underberger D, Steinberg-Koch S, White D, Myasoedova E. Development and Evaluation of a Machine Learning Model for the Early Identification of Psoriatic Arthritis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/development-and-evaluation-of-a-machine-learning-model-for-the-early-identification-of-psoriatic-arthritis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/development-and-evaluation-of-a-machine-learning-model-for-the-early-identification-of-psoriatic-arthritis/