Session Information
Session Type: ACR Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose:
Epigenetics can contribute to the
pathogenesis of rheumatoid arthritis (RA). A DNA methylation signature that
distinguishes RA fibroblast-like synoviocytes (FLS)
from osteoarthritis (OA) FLS has been characterized in previous studies using RA,
OA and normal samples. To confirm the pattern, we evaluated DNA methylation
patterns of an independent group of 19 RA and 5 OA FLS and compared them to
previous findings.
Methods:
Genomic DNA from FLS was isolated
from 19 RA and 5 OA syovia obtained at the time of
total joint replacement. Methylation levels were measured using Illumina HumanMethylation450 chip. Differentially
methylated loci (DMLs) were identified using Welch’s t-test and mapped to gene
promoter regions to define differentially methylated genes (DMGs). Both sets of
DMLs and DMGs were compared to our previously characterized methylation
patterns. To confirm enriched biological pathways, Ingenuity pathway analysis
was applied. Hierarchical clustering and principal component analysis (PCA)
were performed to assess the relationships between 19 RA and 5 OA FLS and also
the combined dataset (30 RA/16 OA).
Results:
2,956 DMLs were identified between
19 RA and 5 OA, and 72.5% were overlapped with DMLs identified in previously
reported data; after mapping DMLs to gene promoter region, 71.5% of 450 DMGs
were overlapped (p-value = 4.26e-284). With established DMLs, distinct
methylation patterns were confirmed between RA and OA. 13 out of 31 significantly
enriched pathways were overlapped with pathways identified previously (p value
< 0.05). Interestingly, “Role of Osteoblasts, Osteoclasts and Chondrocytes
in Rheumatoid Arthritis” ranked No.1 among them. Additional overlapped pathways
such as “Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid
Arthritis”, “Atherosclerosis Signaling”, “Leukocyte
Extravasation Signaling”, and “Angiopoietin
Signaling” suggested the DMGs contribute to inflammation, immunity and joint
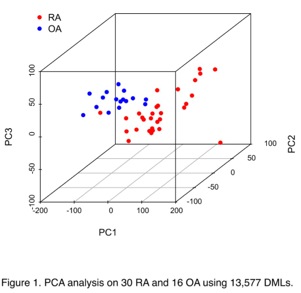
destruction. With the combined dataset (30 RA/16 OA), 13,577 DMLs in 1,714 DMGs
were found, and 43 out of 66 significantly enriched pathways overlapped with
previous data, especially in pathways involving inflammation and immune
responses. Using hierarchical clustering and PCA, the 30 RA and 16 OA segregated
into two groups (Figure 1).
Conclusion:
The significantly overlapped DMLs/DMGs
between the new and previously reported FLS lines confirmed the consistency of DNA
methylation signatures and FLS imprinting. In addition, overlapping enriched
biological pathways suggest that the methylation patterns might contribute to
the pathogenesis of RA. With the expanded dataset, DNA methylation signatures
of RA become more powerful and can provide insights into pathogenesis of the
disease and identify potential therapeutic targets.
To cite this abstract in AMA style:
Ai R, Hammaker D, Wang W, Firestein GS. Confirmatory Analysis of Methylome Signatures in Rheumatoid Arthritis Using an Independent Dataset [abstract]. Arthritis Rheumatol. 2015; 67 (suppl 10). https://acrabstracts.org/abstract/confirmatory-analysis-of-methylome-signatures-in-rheumatoid-arthritis-using-an-independent-dataset/. Accessed .« Back to 2015 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/confirmatory-analysis-of-methylome-signatures-in-rheumatoid-arthritis-using-an-independent-dataset/