Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose: A major challenge in human genetics is to devise a systematic strategy to integrate disease-associated variants with diverse genomic and biological datasets to provide insight into disease pathogenesis and guide drug discovery. Here, we demonstrate one such strategy for a common autoimmune disease with no known cure, rheumatoid arthritis (RA).
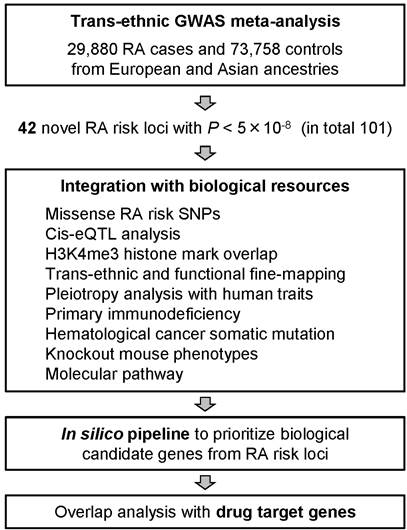
Methods: We performed a trans-ethnic genome-wide association study (GWAS) and in silico and de novo replication studies in a total of >100,000 subjects of European and Asian ancestries (29,880 RA cases and 73,758 controls), by assessing ~10 million autosomal and X-chromosomal single nucleotide polymorphisms (SNPs). Using the RA risk loci obtained from the GWAS meta-analysis, we conducted an integrative analysis with a variety of biological resources: functional annotation of SNPs (missense or non-coding); cis-eQTL analysis for peripheral blood mononuclear cell (PBMC) / T cells / monocytes; H3K4me3 histone peak overlap; trans-ethnic and functional fine-mapping of candidate causal alleles; pleiotropy analysis with other human complex traits; and others. Armed with these biological insights or RA, we constructed in silico pipeline to prioritize biological candidate genes from the RA risk loci, and evaluated their connections to target genes for approved RA drugs.
Results: We discovered 42 novel RA risk loci at a genome-wide level of significance (P < 5x10-8), bringing the total to 101. The common alleles at these RA risk loci revealed: a shared genetic architecture among individuals of European and Asian ancestry; most risk alleles alter gene expression with fewer alleles altering protein structures; two-thirds of loci had pleiotropic effects on other traits, especially disorders of the immune system and inflammatory biomarkers; an overlap with genes that contribute to human primary immunodeficiency (PID) and hematological cancer somatic mutations; and specific cell types (e.g. overlap with H3K4me3 peaks in CD4+ regulatory T cells) and molecular pathways (e.g., T cell, B cell, cytokine signaling) that contribute to RA pathogenesis. We also demonstrated that biological candidate RA risk genes were significantly enriched in overlap with genes that are the target of approved therapies for RA (e.g., TNF and IL6R), and further suggested that drugs approved for other indications may be repurposed for the treatment of RA (e.g., CDK4/CDK6 inhibitors used in cancers).
Conclusion: This comprehensive genetic study sheds light on fundamental pathways and genes that contribute to RA pathogenesis, and provides empirical evidence that the genetics of RA can provide important information for drug discovery efforts.
Disclosure:
Y. Okada,
None;
D. Wu,
None;
C. Terao,
None;
K. Ikari,
None;
Y. Kochi,
None;
K. Ohmura,
None;
A. Suzuki,
None;
H. Yamanaka,
None;
J. C. Denny,
None;
J. D. Greenberg,
None;
R. R. Graham,
Genentech and Biogen IDEC Inc.,
3;
M. A. Brown,
None;
S. C. Bae,
None;
J. Worthington,
None;
L. Padyukov,
None;
L. Klareskog,
No own commercial interests,
2;
P. K. Gregersen,
None;
P. M. Visscher,
None;
K. A. Siminovitch,
None;
R. M. Plenge,
Corrona,
5,
Ignyta,
4.
« Back to 2013 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/biological-insights-from-genetics-of-rheumatoid-arthritis-contribute-to-drug-discovery/