Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic inflammatory diseases (SIDs) are characterized by non-infectious multisystem inflammation. Genetic panels only diagnose 25% of suspected SID patients, due to the clinical and genetic heterogeneity of SIDs. We hypothesize that integration of clinical and laboratory data will improve the diagnosis and evaluation of SIDs by creating shared biologically-based patient phenotypes. We aimed to identify patient clusters with shared clinical and laboratory features within a heterogeneous SID cohort, using similarity network fusion (SNF), a method for multifactorial data integration.
Methods: We included pediatric genetically undiagnosed SID patients with clinical, laboratory, and whole-exome sequencing data. Disease features were divided into clinical and laboratory data sets, weighted by missingness and the number of variables in each data set, then input into SNF. Fisher’s exact test (Bonferroni corrected P< 0.0004) identified variables with different distributions between clusters. We validated our model with 2 simulations (1000 simulated SNF each). Simulation 1: each iteration randomly re-distributed relevant variables into 2 simulated data sets, performed SNF and identified variables with different distributions between simulated clusters. Simulation 2: each iteration randomly subsampled 70% of our cohort into a simulated data set, performed SNF, and determined the percent of patients in the simulated data set that had identical clustering to our original clusters.
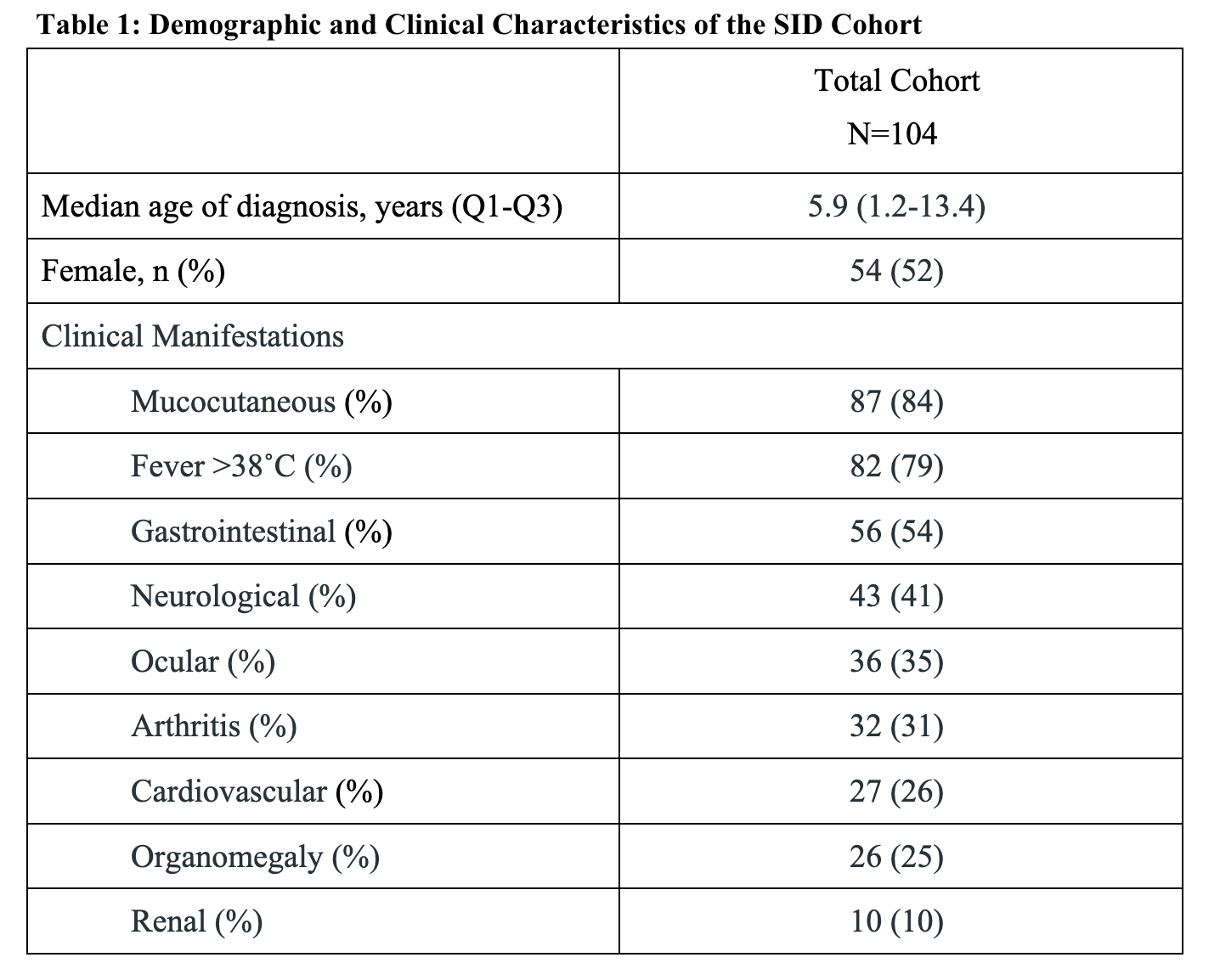
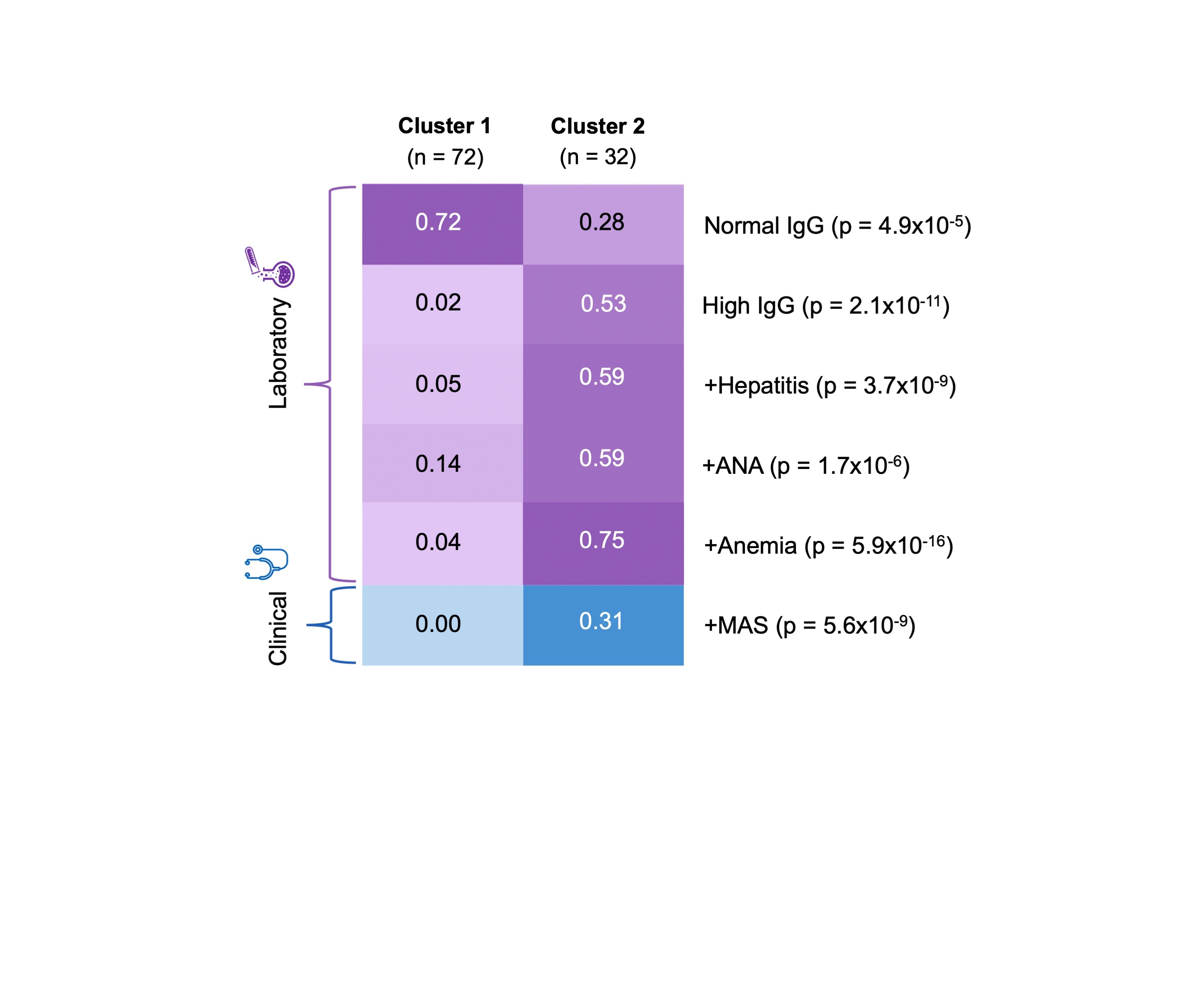
Results: Our study included 104 patients with undifferentiated SID (table 1). SNF revealed 2 clusters in our cohort. Cluster 1 (n=72) had a median age of symptom onset of 5.7 years and cluster 2 (n=32), 6 years. Cluster 2 included patients with more severe SID features compared to cluster 1. Specifically, elevated levels of IgG antibodies and the presence of hepatitis, anti-nuclear antibodies, anemia, and macrophage activation syndrome characterized cluster 2 and were normal/absent in cluster 1 (P< 1×10-7; figure 1). The majority of variables that differentiated the clusters were laboratory features. Simulation 1 demonstrated that the same features were consistently significant in defining clustering ( >946 times out of 1000). Simulation 2 illustrated that 95% of cluster 1 patients (69/72) and 82% of cluster 2 patients (26/32), consistently clustered together over 1000 SNF iterations.
Conclusion: SNF identified 2 relatively homogenous and robust patient clusters from a heterogeneous SID cohort. Simulations showed that 89% of the patients consistently clustered together and that laboratory variables primarily determined these clusters. Future work will incorporate genetic data into SNF to identify shared genetic and inflammatory pathways within clusters.
To cite this abstract in AMA style:
Chan N, Rueppell A, Dominguez D, Appleton T, Batthish M, Berard R, Cellucci T, Demirkaya E, Diebold M, Heale L, Kannu P, Levy D, Park J, Proulx-Gauthier J, Punnett A, Roth J, Schneider R, Spiegel L, Yeung R, An J, Jain A, Couse M, Dissanayake D, Laxer R, Erdman L, Hiraki L. Applying Similarity Network Fusion to Identify Patient Clusters for People with Systemic Inflammatory Disease [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/applying-similarity-network-fusion-to-identify-patient-clusters-for-people-with-systemic-inflammatory-disease/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/applying-similarity-network-fusion-to-identify-patient-clusters-for-people-with-systemic-inflammatory-disease/