Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: There is a need to design models based on machine learning that are capable of predicting the risk of rapid pain progression of knee OA (RAPPKOA) in order to establish personalized treatment patterns. Our goal is to combine clinical and molecular variables to design a model capable of predicting rapid pain progression using well-characterized prospective cohorts.
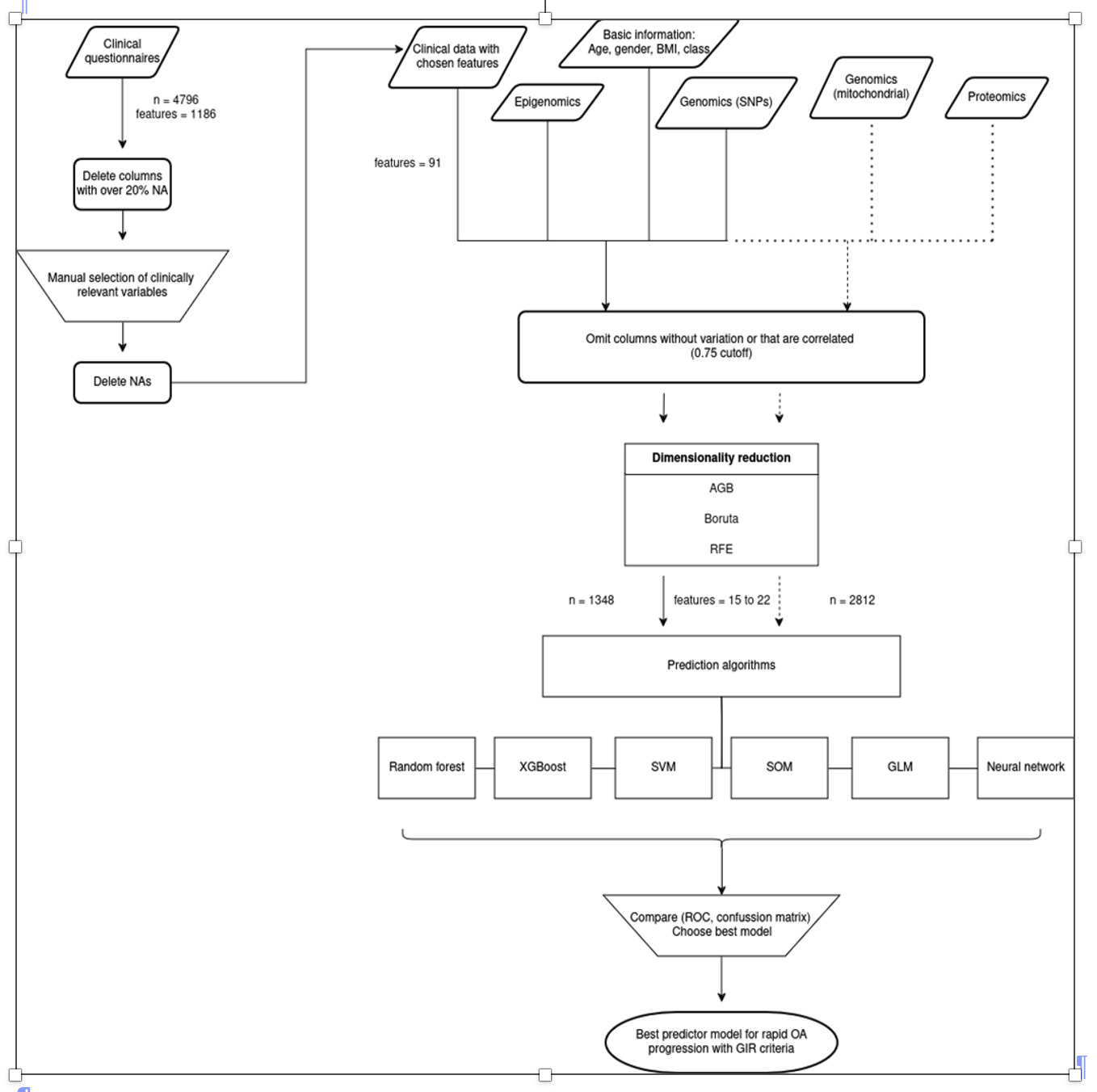
Methods: Rapid pain progressors of knee OA (RAPPKOA) were defined as patients whose knee pain has increased by at least minimal clinically important difference (5 WOMAC pain points on a 0–100 scale) per year and is substantial at the end of a 48-month follow-up period (at least 40 WOMAC pain points), or those with substantial pain (at least 40 WOMAC pain points) that was sustained both at the start and the end of the period. To construct the models, we used different clinical, genomic, proteomic and epigenetic data that were subsequently filtered using a feature selection strategy. As part of the genomic variables, we have included significant SNPs (< 10-7) resulting from the GWAS performed in the OAI database and in PROCOAC. Polygene Risk Score (PRS) was defined for RAPPOA phenotype. Different machine learning algorithms were tested to find the best one for prediction. Datasets were divided into 75% for training and 25% for test. A validation of the model in PROCOAC was performed using those variables that were common between both cohorts. A flowchart is described in Figure 1.
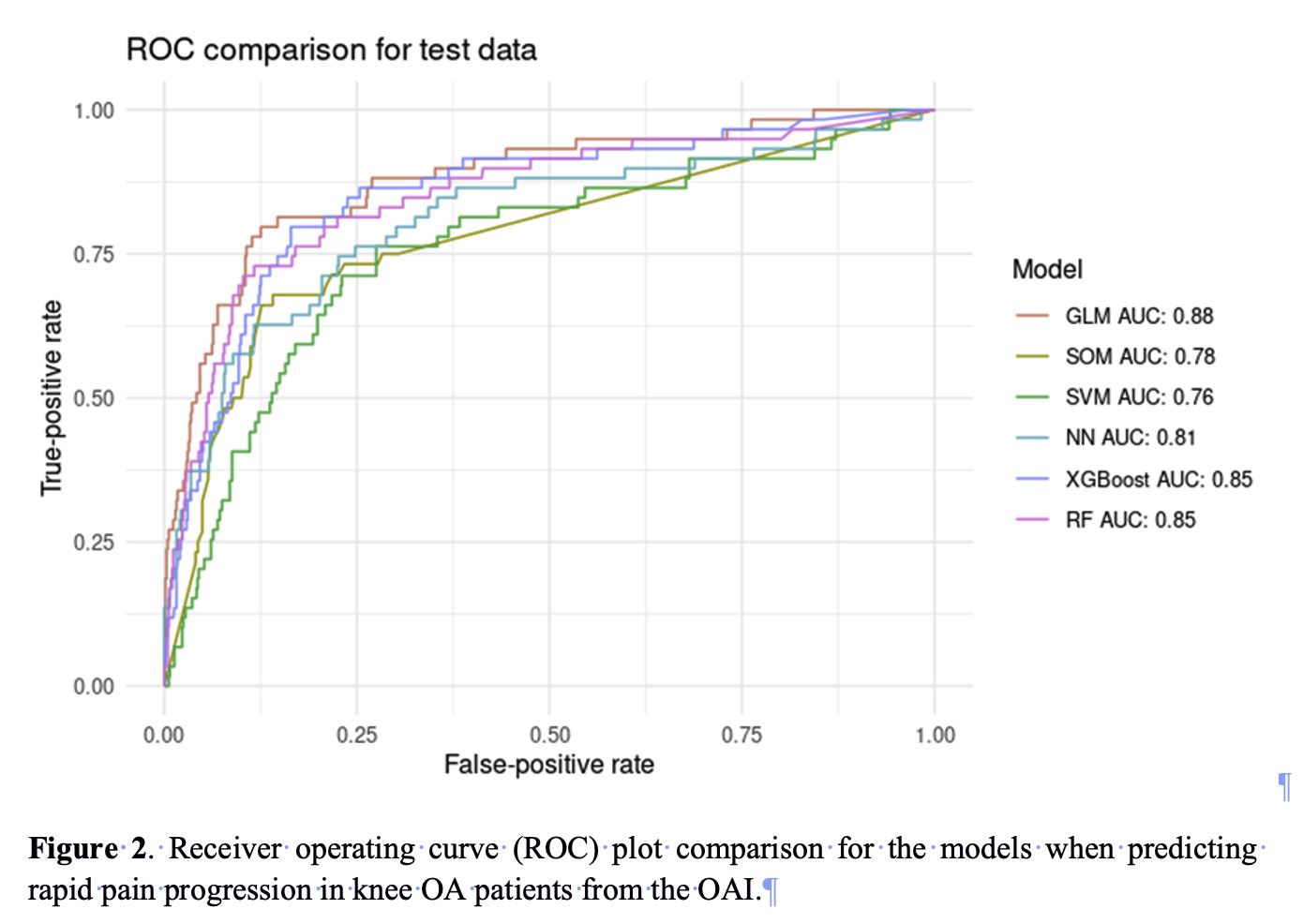
Results: The discovery cohort (OAI) consisted of 472 rapid pain progressors, whereas the validation cohort (PROCOAC) consisted of 131 rapid pain progressors. The predictive model included up to 16 variables, highlighting KOOS sub-scores, body mass index, symptomatic knee OA at baseline, WOMAC stiffness score, knee pain, use of pain medications, lower adherence to Mediterranean diet as well as three nuclear SNPs (rs111308634, rs35940114 and rs114970041). The best machine learning algorithm was GLMnet, showing an area under the curve (AUC) of 0.88 and a sensitivity and specificity values of 81% (Figure 2).
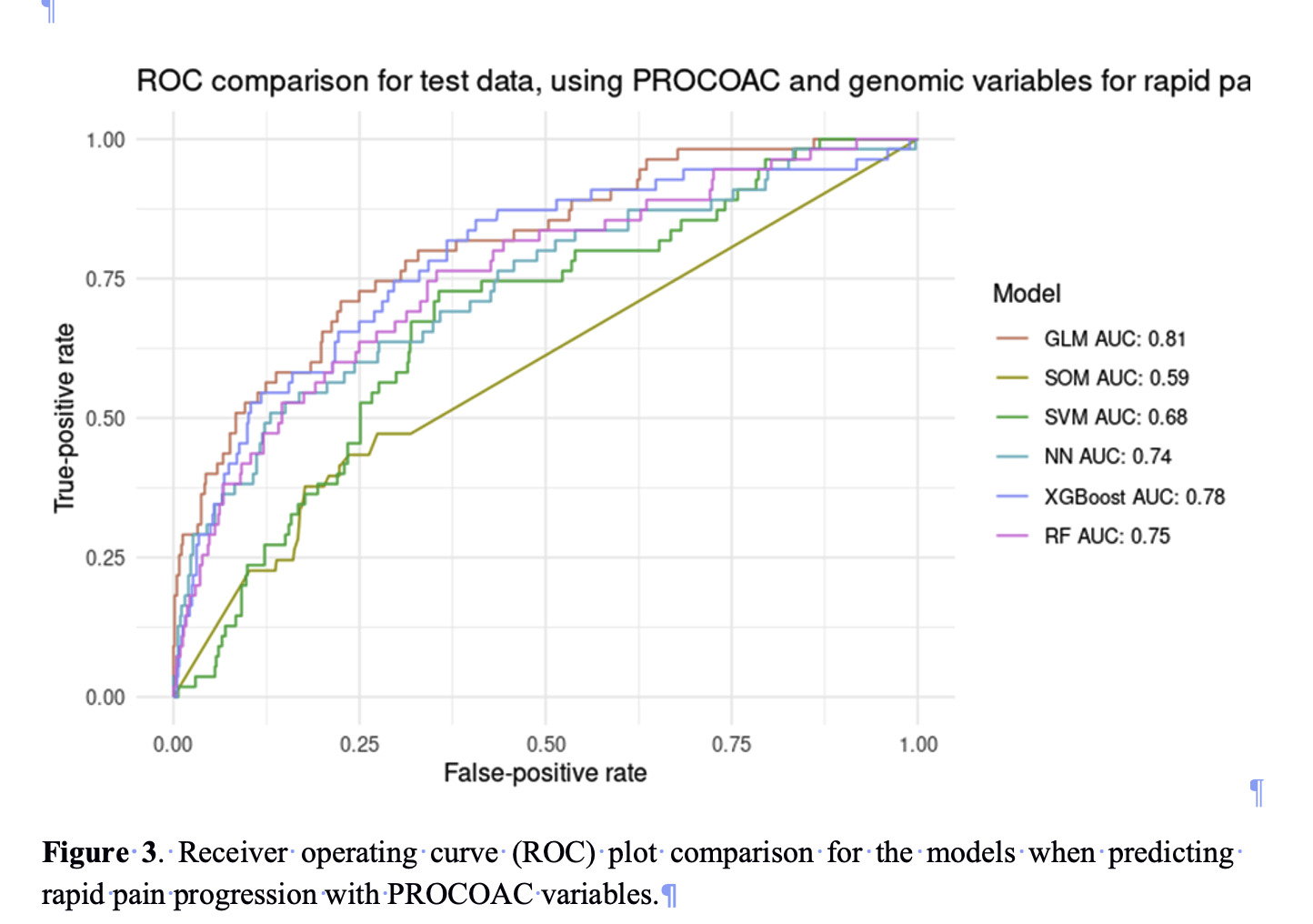
Further validation was pursued by attempting to test the models on PROCOAC cohort, but this cohort did not contain the same variables as the original OAI. For this reason, new models were calculated that only used variables common between both OAI and the PROCOAC cohorts. The models trained with these limited variables performed slightly worse overall, being GLMnet still the best model, showing and AUC of 0.81 and 70% both specificity and sensitivity (Figure 3).
Conclusion: We constructed a series of predictive models based on machine learning that are capable of predicting the risk of rapid pain progression. The design of these models combining different sources of variables are key to deep into the application of precision medicine in the clinical practice.
To cite this abstract in AMA style:
Rego-Perez I, Rodriguez-Valle I, Fernández-Tajes J, Balboa-Barreiro V, Fraga-Seijas C, de andres M, Calamia V, Fernández-Puente P, Veronese N, Ruiz-Romero C, Oreiro N, Blanco f. Application of Machine Learning Methods to Predict the Risk of Rapid Pain Progression in Patients with Knee OA. Study Using Patients from the OAI and PROCOAC [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/application-of-machine-learning-methods-to-predict-the-risk-of-rapid-pain-progression-in-patients-with-knee-oa-study-using-patients-from-the-oai-and-procoac/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/application-of-machine-learning-methods-to-predict-the-risk-of-rapid-pain-progression-in-patients-with-knee-oa-study-using-patients-from-the-oai-and-procoac/