Session Information
Date: Wednesday, November 13, 2019
Title: 6W013: SLE – Clinical VI: Epidemiology, Diagnosis, & Outcomes (2882–2887)
Session Type: ACR Abstract Session
Session Time: 9:00AM-10:30AM
Background/Purpose: Gut microbiota may be relevant in the regulation of immune processes and in the development of systemic autoimmune diseases (SADs). A reduction of microbiota diversity and a selective decrease of commensal pro-regolatory bacteria have been observed in systemic lupus erythematosus (SLE), Sjogren syndrome (SJS) and primary anti-phosholipid syndrome (PAPS). None studies have been performed on undifferentiated connective tissue disease (UCTD) and into an enlarged panel of SADs at the very same time, integrating microbiome data with the plasma metobolomic profile.
Objectives: to perform an integrated investigation of microbiomic and metabolomic profile in patients with SADs and healthy controls (HC) within the PRECISESADs study.
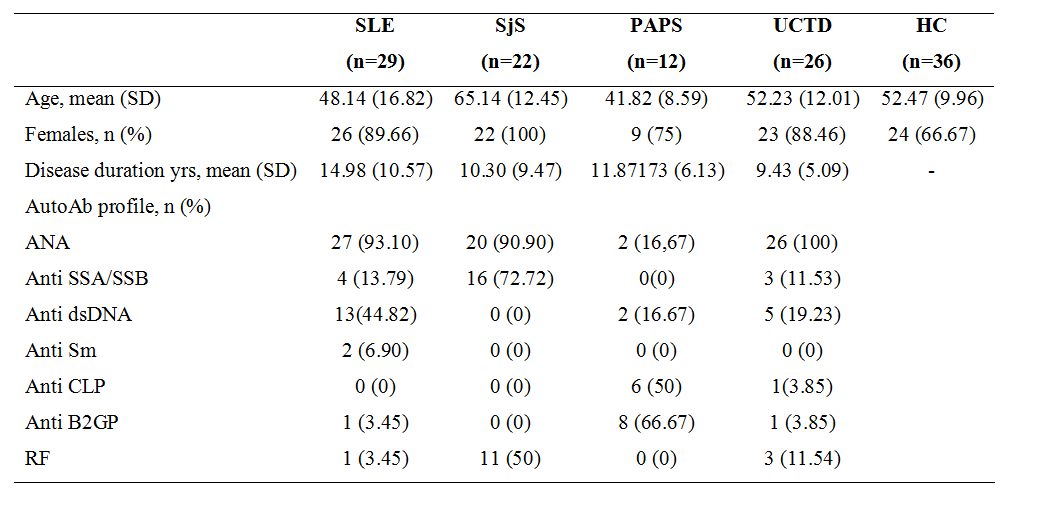
Methods: a total of 125 subjects (29 SLE, 22 SJS, 12 PAPS, 26 UCTD, 36 HC) were enrolled (Table 1). Gut microbiome analysis through 16s-RNA sequencing on stool samples and metabolomic analysis on plasma samples with high-performance liquid chromatography coupled to electrospray ionization and quadrupole time-of-flight mass spectrometry (HPLC-MS-ESI-QTOF) were performed. Data mining algorithms were used to assess a single subset of microbiomic and metabolomic variables capable of to jointly distinguishing the groups of subjects. Microbial and metabolic signals were correlated adjusting for potential confounders (age, therapy) and corrected with stepwise procedure, minP. A Kruskall-Wallis (KW) test with a post hoc Dunn test were used for statistical analysis of microbial genera relative abundance and for metabolic peaks within groups.
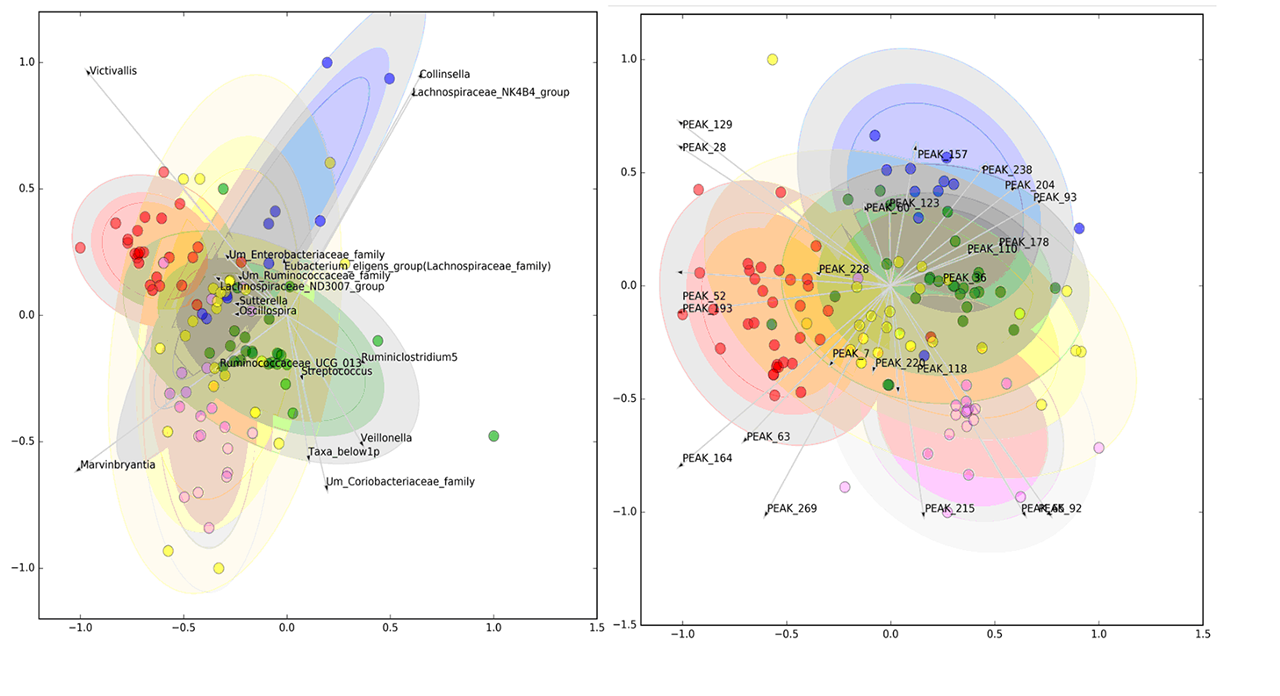
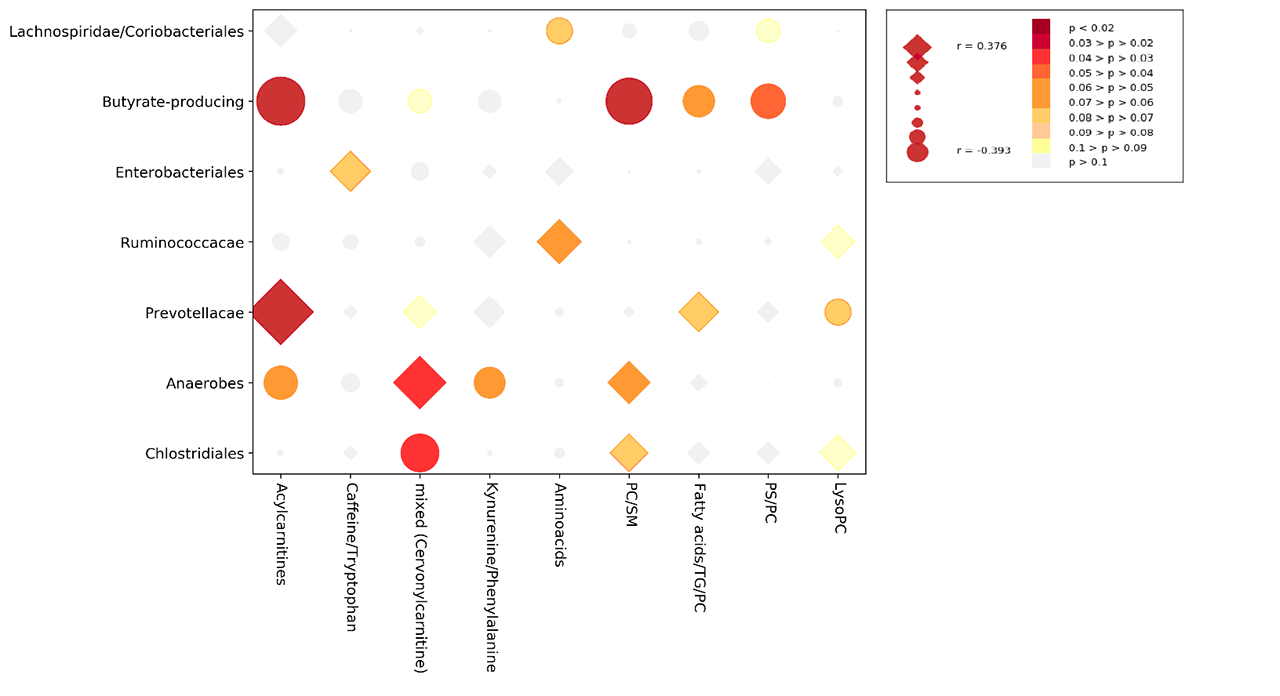
Results: From 130 gut bacterial genera, a subset of 29 was able to jointly distinguish the study groups (AUROC=0.730±0.025). Similarly, of 254 metabolic peaks, 41 were able to discriminate study groups (AUROC=0.748±0.021). In both models, HC were well separated from SADs, while UCTD largely overlapped with other disease groups (Figure 1). The relative abundance of pro-tolerogenic bacterial strains such as Lachnospiraceae was reduced in SADs (p=1.77E-06) while an increase of pro-inflammatory genera (Collinsella, Streptococcus, Clostridiales with p< 0.001) was observed. Metabolic alterations included mostly members of the carnitine family and phospholipids. Microbiomic genera significantly correlated with metabolic peaks as shown in Figure 2 and Acylcarnitines were directly correlated with a Prevotella-enriched cluster and inversely correlated with a butyrate-producing bacteria-enriched cluster. The latter was also inversely correlated with a phospholipid-enriched cluster.
Conclusion: A common microbiomic and metabolomic profile jointly distinguish SLE, PAPS, SJS, UCTD and HC. SADs are characterized by a reduction of pro-tolerogenic bacterial genera and the presence of correlations between microbial genera and metabolites support the hypothesis of an interaction between intestinal microbiota and metabolic functions.
This work was supported by EU/EFPIA/Innovative Medicines Initiative Joint Undertaking PRECISESADS grant No. 115565.
To cite this abstract in AMA style:
Bellocchi C, Fernández-Ochoa �, Montanelli G, Vigone B, Santaniello A, Quirantes-Piné R, Borrás-Linares I, Gerosa M, Artusi C, Gualtierotti R, Segura-Carrettero A, Alarcón-Riquelme M, Beretta L. An Integrated Gut Microbiomic and Plasma Metabolomic Analysis in Patients with Four Systemic Autoimmune Diseases [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/an-integrated-gut-microbiomic-and-plasma-metabolomic-analysis-in-patients-with-four-systemic-autoimmune-diseases/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/an-integrated-gut-microbiomic-and-plasma-metabolomic-analysis-in-patients-with-four-systemic-autoimmune-diseases/