Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: The NT5E gene, encoding Ecto-5′-Nucleotidase/CD73, is expressed on the surface of immune system cells and is critical for converting AMP/IMP to adenosine/inosine, important immunosuppressive purine nucleosides. The p.Met379Thr (rs2229524 T to C) substitution in the NT5E gene has been linked to systemic lupus erythematosus (SLE) in individuals of European ancestry,1,2 and our recent data support impaired adenosine-mediated signaling and augmented expression of inflammatory mediators in SLE patients bearing the minor allele NT5E variant (abstract submitted). The clinical significance of this variant is supported by its association with hypertension and pericarditis in our SLE cohort. Given this background, we aimed to characterize peripheral blood NT5E expression based on genotype in our SLE patients and to define the major cell types expressing NT5E in a single-cell RNA-seq skin tissue dataset derived from SLE patients with skin manifestations.
Methods: We genotyped 267 SLE patients using Sanger sequencing and used the 1000 Genomes Project population as a reference control. Minor allele frequencies were calculated using snpReady, and association analysis was performed with SNPassoc statistical tools. Gene transcript levels in PBMCs were assessed in 122 bulk RNA-seq datasets from selected patients. We also examined NT5E levels in dermal skin cell populations using a single-cell RNA-seq dataset (GSE179633) from lesional skin biopsies of 6 patients with SLE and 4 healthy controls (HC).
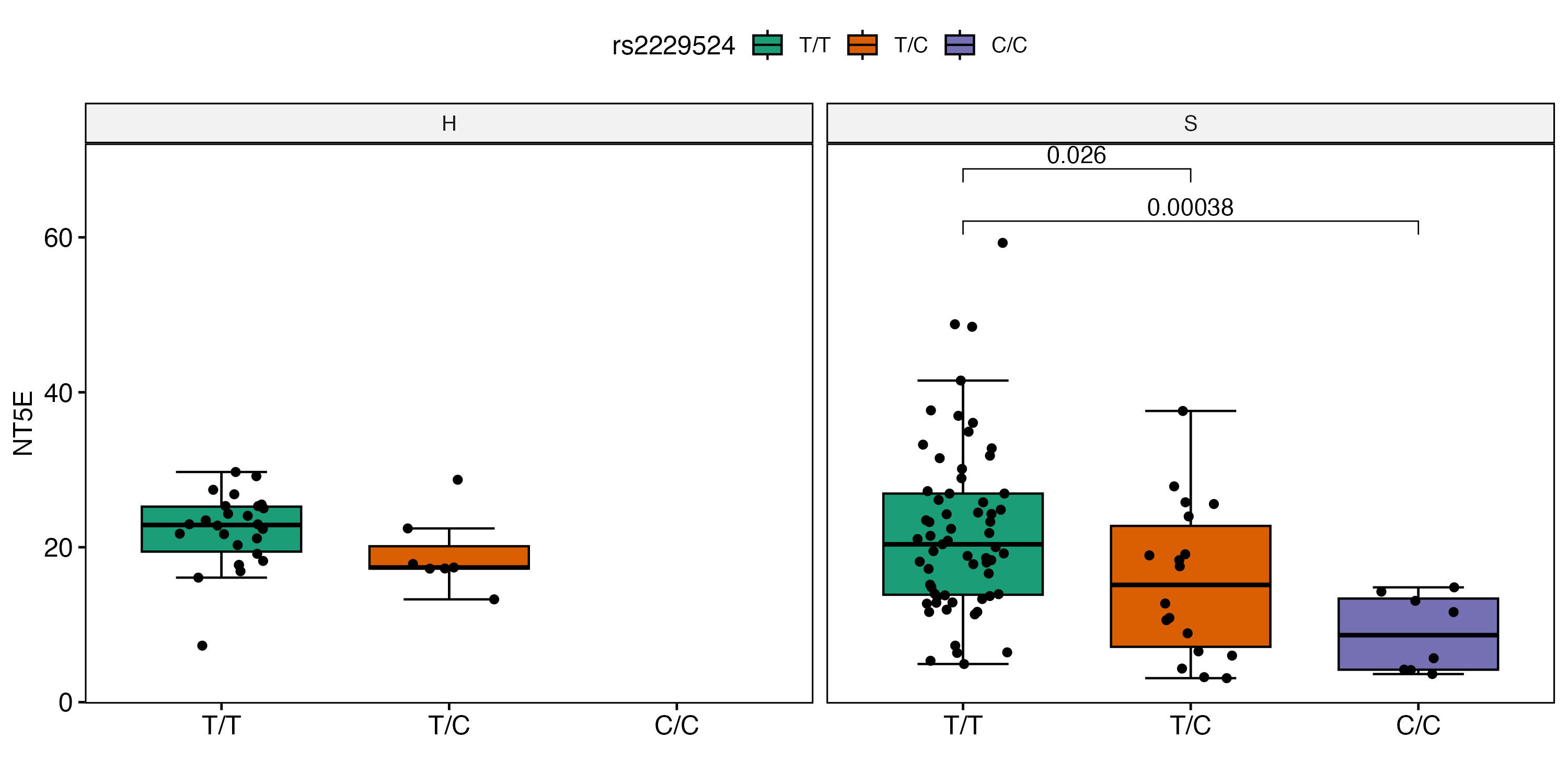
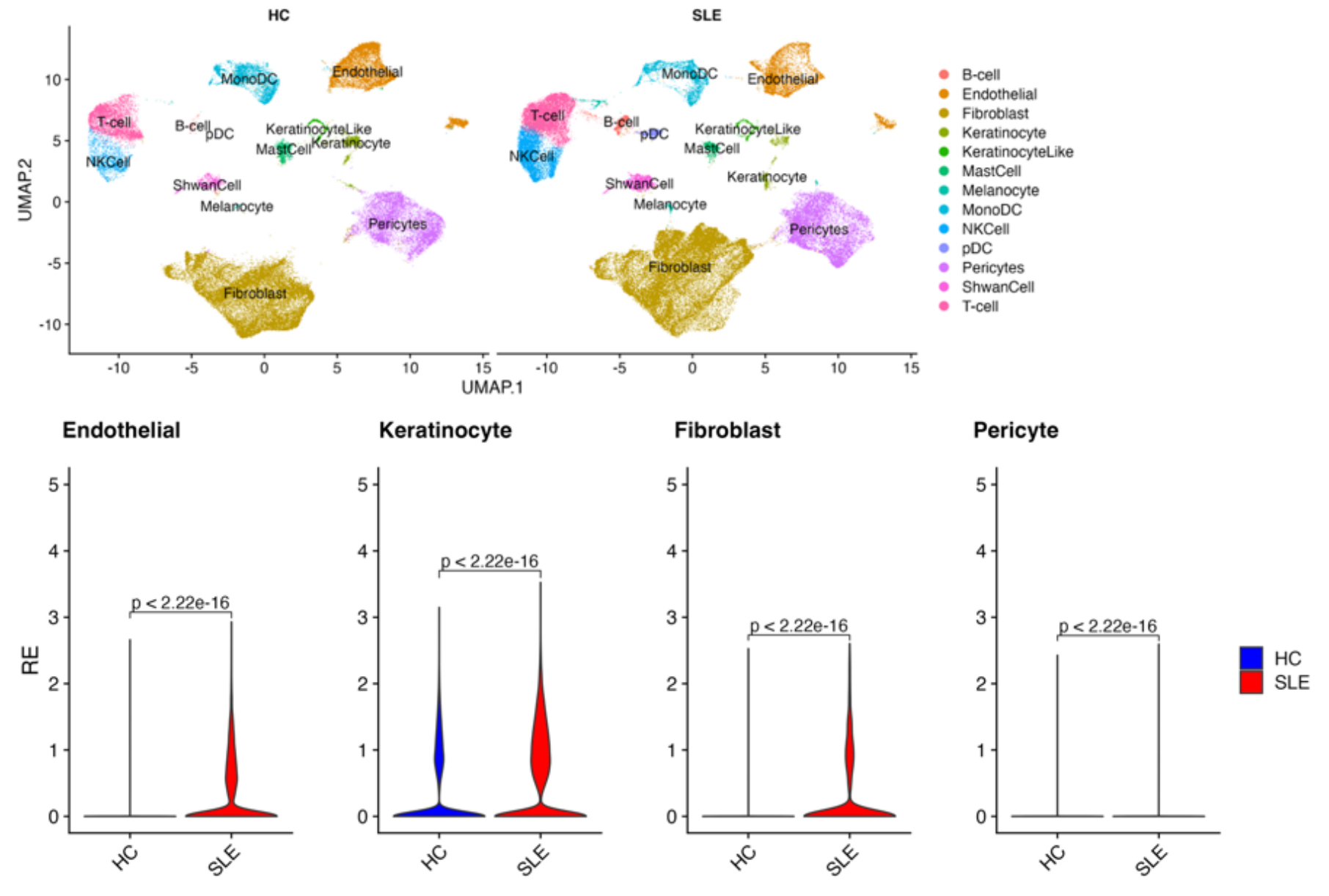
Results: To relate gene expression to rs2229524 genotype, we investigated expression of NT5E in RNA-seq datasets from patients in our HSS SLE cohort defined by NT5E genotype. The rs2229524 minor allele variant was associated with decreased NT5E transcript levels in PBMCs of SLE patients (T/C, p=0.026; C/C, p=0.00038) in comparison to those expressing the common allele (T/T, Figure 1). To gain insight into potential mechanisms that might account for association of the minor NT5E allele with vascular disease, we leveraged single-cell RNA-seq data from SLE patients with skin involvement to identify specific cell populations demonstrating altered NT5E expression in dermal skin. We observed a significant increase in NT5E transcript levels in endothelial cells, pericytes, keratinocytes and fibroblasts from SLE skin (Figure 2).
Conclusion: These data extend our observations of altered NT5E expression and adenosine pathway signaling in SLE patients and association of the minor allele NT5E variant with a history of hypertension in our patient cohort. We document decreased expression of NT5E transcripts in PBMC of patients with the minor allele and a predominance of NT5E expression on non-immune cells in skin from SLE patients with skin involvement. Overall, the aberrant expression of NT5E in SLE blood and tissue, with increased expression on endothelial cells and pericytes, may point to a pathologic role for vascular cells in the increased risk of hypertension based on NT5E genotype.
1. Langefeld CD et al. Nat Commun. 2017 Jul 17;8:16021.
2. Owen KA et al. Am J Hum Genet. 2020 Nov 5;107(5):864-881.
To cite this abstract in AMA style:
Olferiev M, Owen K, Lipsky P, Crow M. Altered Expression of Ecto-5′-Nucleotidase (NT5E) in SLE Patients Based on Disease-associated Genotype [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/altered-expression-of-ecto-5-nucleotidase-nt5e-in-sle-patients-based-on-disease-associated-genotype/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/altered-expression-of-ecto-5-nucleotidase-nt5e-in-sle-patients-based-on-disease-associated-genotype/