Session Information
Session Type: Poster Session (Tuesday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Genetic studies of common variants in scleroderma (systemic sclerosis, SSc) have identified several susceptibility loci increasing SSc risk. Most of these common variants are in the non-coding region and thus have a relatively minor effect size and are not able to fully account for the heritability of SSc. The risk accorded by rare and functional variants is presumed to be higher and could be part of the missing heritability.
Methods: We selected all SSc-associated loci at p-value< 0.00001 from the GWAS catalog, an NHGRI-EBI catalog of published genome-wide association studies and genes identified in candidate gene studies that were independently replicated. 26 genes were selected and sequenced using next generation sequencing technology in 379 SSc patients and 411 unaffected controls of Africa. Variants were selected for testing if they were missense, splice site or InDels and based on CADD score < 15 for synonymous and UTR variants. Association testing was performed using sequence kernel association test (SKAT). Significance threshold for p-value (P) was set at P< 0.002 after Bonferroni’s multiple testing correction.
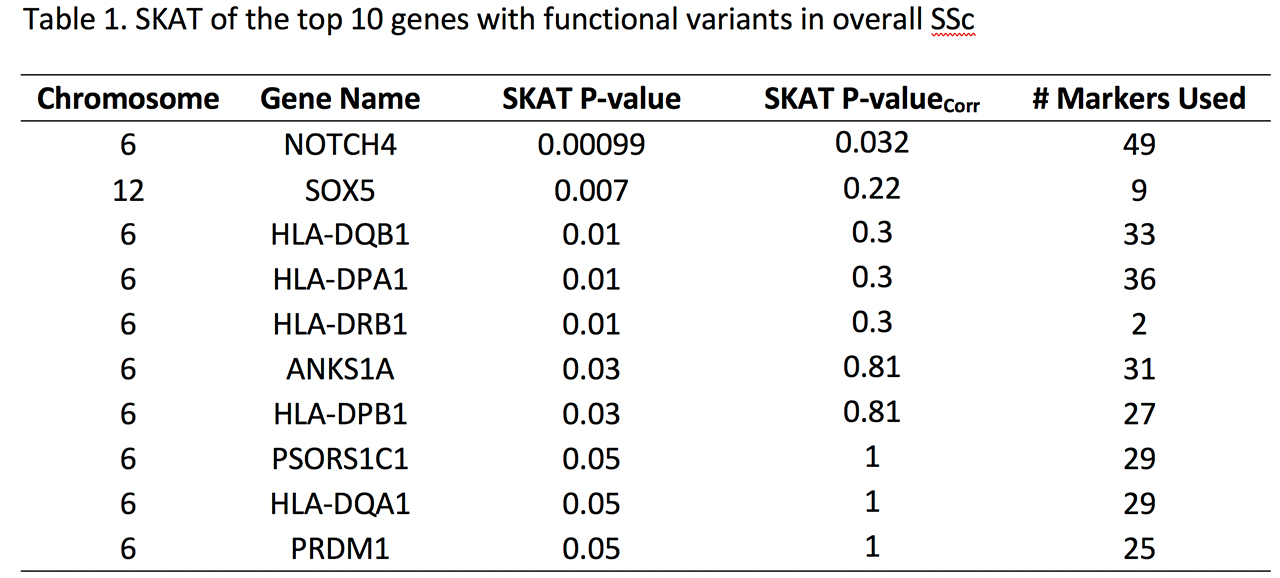
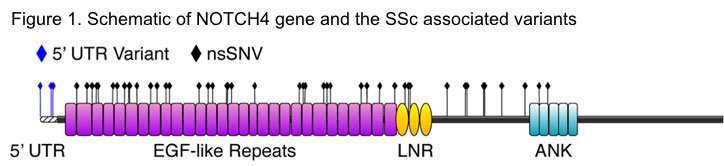
Results: After multiple testing correction only NOTCH4 (Neurogenic Locus Notch Homolog Protein 4) gene was statistically significantly associated with SSc (P=0.00099, Pcorr=0.03) with an increased burden of functional variants (Table1). We used CMC test to confirm the NOTCH4 association and the P remained significant (CMC P=0.01, 100000 permutations). Majority of the variants were present on the extracellular domain of NOTCH4 (Figure 1). On comparing the phenotypic subsets of SSc, the NOTCH4 association was strongest in the diffuse skin subset (P=0.001) and interstitial lung disease subset of SSc (P=0.0003).
Conclusion: Aggregation of functional variants in genes previously implicated with SSc provides new approaches to understand SSc pathogenesis. Understanding the functional role of these NOTCH4 functional variants will increase our understanding of SSc and SSc-associated interstitial lung disease.
To cite this abstract in AMA style:
Gourh P, Safran S, Alexander T, Boyden S, Shah A, Mayes M, Doumatey A, Bentley A, Shriner D, Domsic R, Medsger Jr T, Ramos P, Silver R, Steen V, Varga J, Hsu V, Saketkoo L, Schiopu E, Khanna D, Gordon J, Kron B, Criswell L, Gladue H, Derk C, Bernstein E, Bridges S, Shanmugam V, Kolstad K, Chung L, Kafaja S, Jan R, Trojanowski M, Goldberg A, Korman B, Steinbach P, Chandrasekharappa S, Mullikin J, Adeyemo A, Rotimi C, Wigley F, Kastner D, Boin F, Remmers E. Aggregation of Functional Variants in NOTCH4 Gene Increases SSc Risk [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/aggregation-of-functional-variants-in-notch4-gene-increases-ssc-risk/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/aggregation-of-functional-variants-in-notch4-gene-increases-ssc-risk/