Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic Lupus Erythematosus (SLE) is a complex autoimmune disorder associated with increased cardiovascular risk. Platelet activity is increased in SLE and may mediate, in part, the excess cardiovascular risk observed in patients with SLE. Our group previously developed a platelet gene expression score associated with SLE prevalence and disease activity. This study aims to validate this score and explore its association with platelet activity and vascular health.
Methods: Participants from the NYU Lupus Cohort were enrolled and were assessed for disease activity. Our previously defined lupus prevalence signature, based on 31 genes, was compared between groups and evaluated using area under the curve (AUC). The SLAP-GES (SLE Activity Platelet – Gene Expression Signature) score, incorporating 548 genes, was calculated in each participant using singscore. To investigate the clinical relevance of SLAP-GES, we assessed its association with platelet aggregation (light transmission aggregometery and leukocyte/monocyte/neutrophil-platelet aggregation) and both micro- and macro-vascular health. For micro-vascular, the glycocalyx was examined with sublingual sidestream dark field microscopy (microvascular percent RBC filling and perfused boundary region of vessels between 5 and 25 µm in diameter (PBR 5-25). Macrovascular health was assessed via brachial artery reactivity testing before (femoral mediated dilation, FMD) and after sublingual nitroglycerin (NMD).
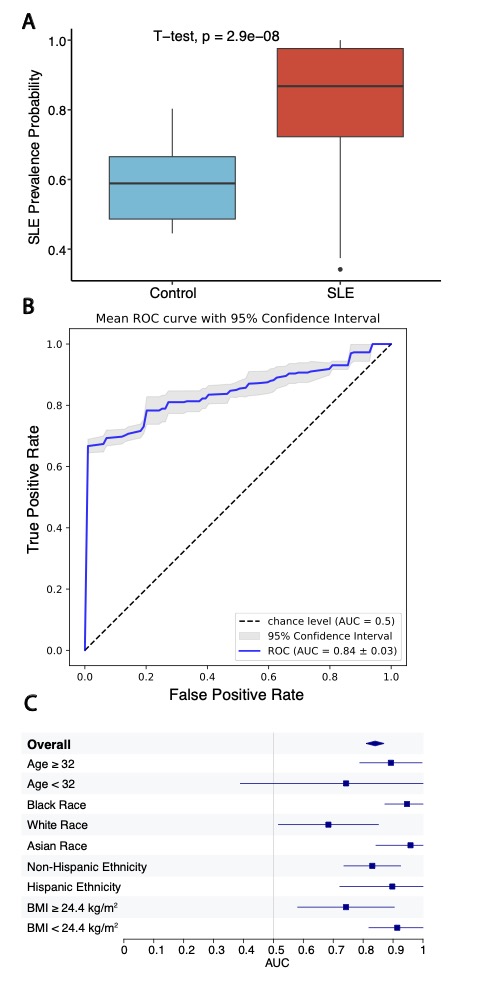
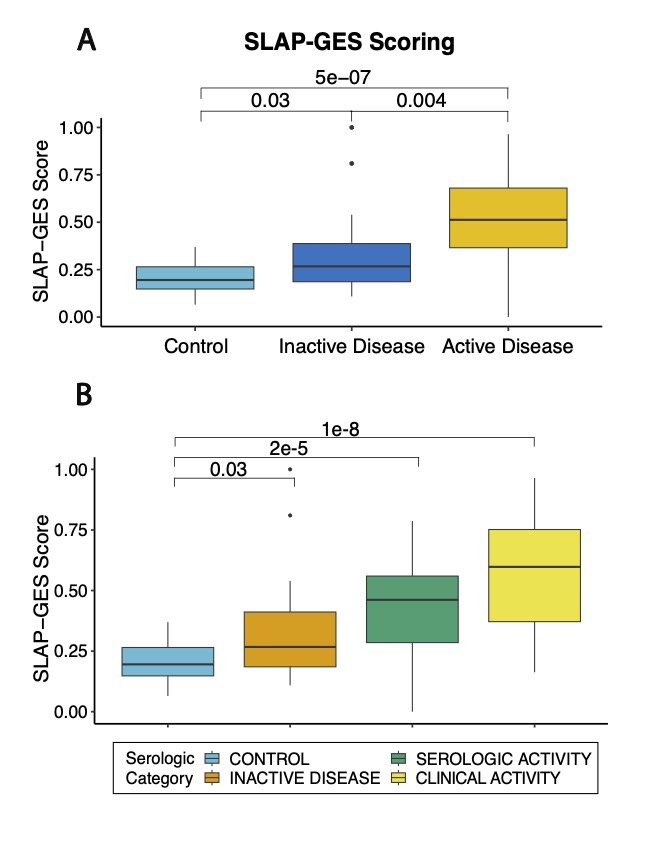
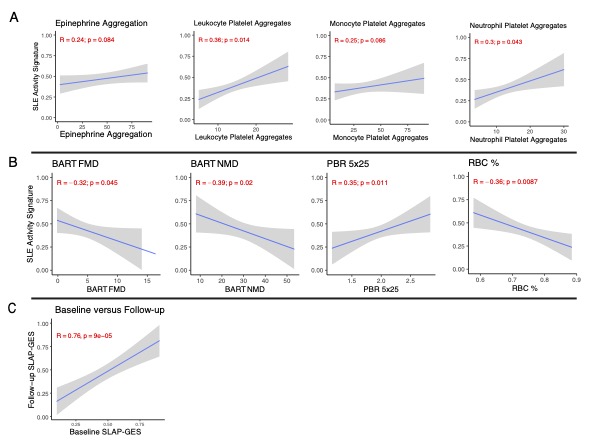
Results: RNA-seq was performed on isolated platelets from an independent cohort of 71 female SLE patients (mean age 38, 42% Black, mean SLEDAI 3.1) and 18 healthy female controls (mean age 26, 56% Black). Transcriptomic analysis identified 488 differentially expressed genes (padj < 0.05), with significant enrichment in pathways related to viral defense (NES 2.42, padj = 1.2e-8) and interferon-mediated signaling (NES 2.20, padj = 1.2e-8). Our previously defined lupus prevalence signature was higher in participants with SLE (Fig 1A) and effectively discriminated SLE patients from controls in this independent cohort (AUC = 0.84; Fig 1B). The prevalence signature remained robust when stratified by age, race, ethnicity, and body mass index (Fig 1C). For each participant, we calculated the SLE Activity Platelet – Gene Expression Signature (SLAP-GES). SLAP-GES was associated with SLEDAI (0, 0.34 ± 0.24; 1-4, 0.43 ± 0.21; >4, 0.58 ± 0.21, P< 0.001, Fig 2A) and clinical domain (Fig 2B). SLAP-GES was independent of cardiovascular risk factors, including increasing age, body mass index, and smoking status. While SLAP-GES was not associated platelet count or size, it was associated with increased platelet aggregation (Fig 3A) and both micro- and macro-vascular health (Fig 3B). Finally, we tested the signature in a subset of 13 SLE participants with transcriptomics measured at two timepoints (median 4 months apart), and demonstrated a significant correlation between timepoints (r= 0.76, p = 9e−05; Figure 3B).
Conclusion: This study validates the use of platelet transcriptomics to discriminate SLE prevalence and SLE disease activity. In addition, we demonstrate the association between platelet gene expression and cardiovascular risk in SLE patients.
To cite this abstract in AMA style:
Muller M, Luttrell-Williams E, Rosmann H, Ruggles K, Buyon J, Berger J. A Transcriptomic Lupus Activity Signature Associates with Cardiovascular Function [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/a-transcriptomic-lupus-activity-signature-associates-with-cardiovascular-function/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-transcriptomic-lupus-activity-signature-associates-with-cardiovascular-function/