Session Information
Session Type: Abstract Session
Session Time: 4:45PM-5:00PM
Background/Purpose: Severe coronavirus disease 2019 (COVID-19) is associated with a broad range of clinical conditions. International efforts have led to the identification of risk alleles associated with severe COVID-19. The objective of this study was to apply the Phenome-Wide Association Study (PheWAS) approach to study conditions sharing risk alleles with severe COVID-19 to inform potential shared pathways across conditions.
Methods: We performed the PheWAS in two mega-biobanks with linked clinical and genomic data: 1) Veteran Affairs (VA) Million Veteran Program (MVP), discovery cohort, 2) United Kingdom Biobank (UKBB), replication cohort. Genetic variants associated with a critical illness (n=48) or hospitalization (n=39) due to COVID-19 were selected from COVID-19 Host Genetics Initiative genome-wide association studies. Phenotypes were defined using a published PheWAS approach using International Classification of Diseases (ICD) codes mapped to clinically relevant groups. Data were extracted on subjects before 2019/pre-COVID-19 to avoid potential confounding. Logistic regression was applied to examine each SNP and phenotypes association and adjusted for sex, age, and the 1st 20 principal components. Ancestry-specific PheWAS was first performed across European (EUR), African (AFR), Hispanic, Asian ancestries and then meta-analyzed using an inverse-variance weighted fixed-effects model.
Results: We studied 455,683 US Veterans from MVP, mean age 68 years, 89% male; 68% EUR, and 19% AFR ancestry. Variants associated with severe COVID-19 were tested for association across 1,559 phenotypes. Genetic variants at the ABO locus (rs550057, rs505922) were associated with the largest number of phenotypes (nrs55057= 53 and nrs505922=61); strongest association with venous embolism (odds ratio (OR)rs550057 1.27, p=5.28 x 10-116), and thrombosis (ORrs505922=1.31, p=3.5 x10-183). Among 67 respiratory conditions tested, only idiopathic pulmonary fibrosis, (ORrs2277732 1.17, p=1.3410-05), and asthma (ORrs143334143 0.94, p=2.31 x10-04), shared variants with severe COVID-19 (Figure). The RAVER1 locus (rs74956615) associated with reduced risk for autoimmune conditions, e.g. psoriasis (OR 0.71, p= 1.53 x10-22), rheumatoid arthritis (OR 0.78, p=1.04 x 10-09) with findings replicating in UKBB (Table). A known functional missense variant (rs34536443, TYK2) in the region had the highest linkage disequilibrium with rs74956615, suggesting the signal stemmed from TYK2. MVP PheWAS results stratified by genetic ancestry did not demonstrate a significant difference in associations across ancestry.
Conclusion: We observed a shared genetic architecture between severe COVID-19 with thrombosis and asthma; this same genetic architecture was also associated with reduced risk of rheumatic conditions, e.g., RA. This divergent association may in part be explained by a TYK2 variant known to impair type 1 interferon signaling, reducing risk for autoimmunity and simultaneously impairing viral host defense. Consideration is needed when targeting pathways that may balance immune tolerance and immunodeficiency to treat COVID-19.
 PheWAS results of candidate SNPs from GWAS of critically ill and hospitalized COVID_19. Significant associations between 48 SNPs from critical ill COVID GWAS (A) and 39 SNPs from hospitalized COVID (C) and EHR-derived phenotypes in the Million Veteran Program. The phenotypes are represented on the x-axis and ordered by broader disease categories. The red line denotes the significance threshold using a false discovery rate of 10% using the Benjamini-Hochberg procedure. The description of phenotypes is highlighted for the associations with FDR < 0.1 and odds ratio < 0.90 or odds ratio > 1.10. B) and D) A heatmap plot of SNPs with at least one significant association (FDR < 0.1). The odds ratio represents the direction of effect disease risk. A red color indicates increased risk, and blue color indicated reduced risk. The results with odds ratio < 0.90 or odds ratio > 1.10 are shown.
PheWAS results of candidate SNPs from GWAS of critically ill and hospitalized COVID_19. Significant associations between 48 SNPs from critical ill COVID GWAS (A) and 39 SNPs from hospitalized COVID (C) and EHR-derived phenotypes in the Million Veteran Program. The phenotypes are represented on the x-axis and ordered by broader disease categories. The red line denotes the significance threshold using a false discovery rate of 10% using the Benjamini-Hochberg procedure. The description of phenotypes is highlighted for the associations with FDR < 0.1 and odds ratio < 0.90 or odds ratio > 1.10. B) and D) A heatmap plot of SNPs with at least one significant association (FDR < 0.1). The odds ratio represents the direction of effect disease risk. A red color indicates increased risk, and blue color indicated reduced risk. The results with odds ratio < 0.90 or odds ratio > 1.10 are shown.
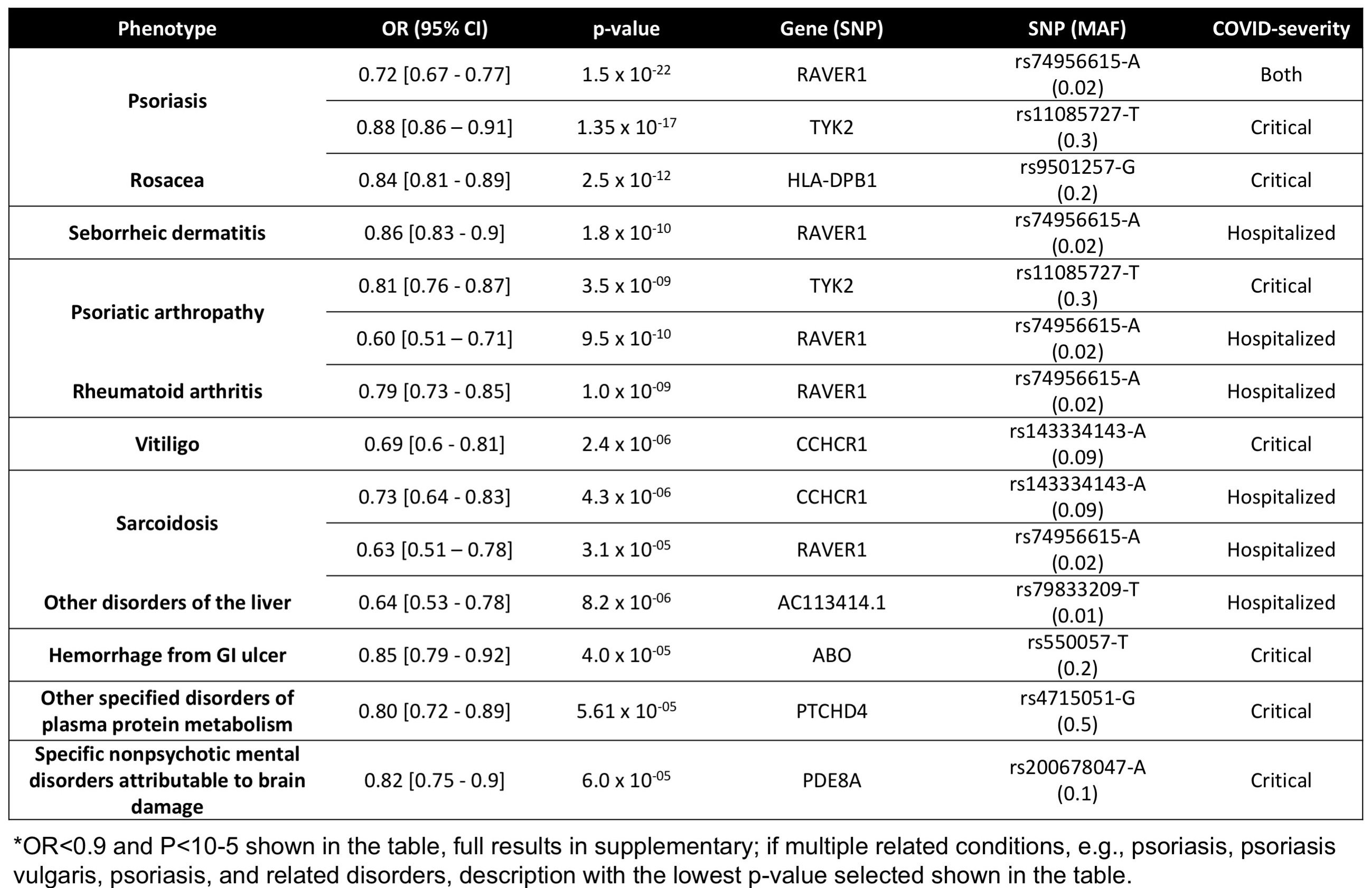 Phenotypes sharing association with variants also associated with severe COVID_19 infection, with reduced odds of disease listed in order of p-value*.
Phenotypes sharing association with variants also associated with severe COVID_19 infection, with reduced odds of disease listed in order of p-value*.
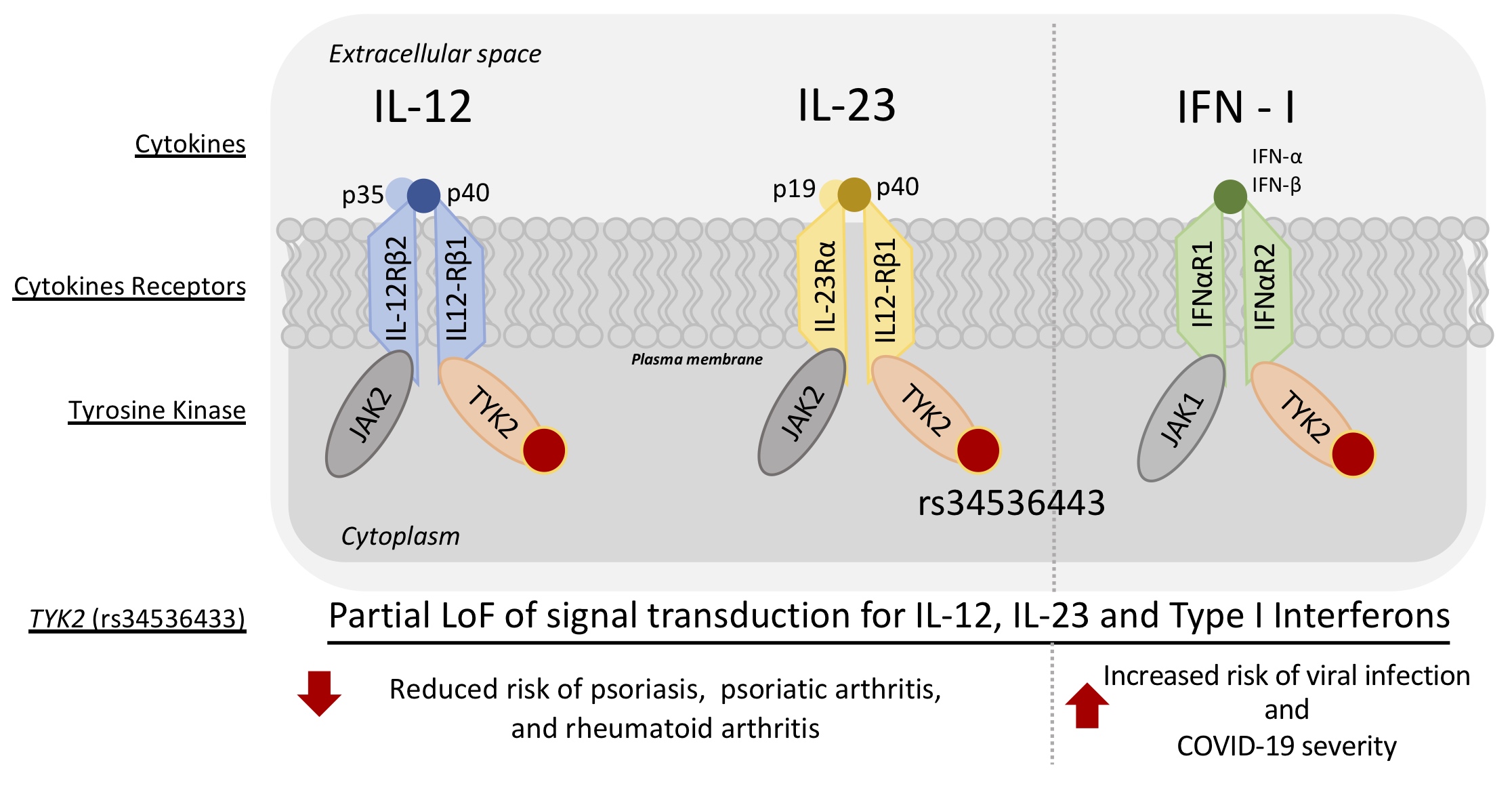 Conceptual model of the relationship between TYK2 (rs34536133) with reduced risk for psoriasis, psoriatic arthritis, rheumatoid arthritis, and simultaneously increased risk of COVID_19 severity.
Conceptual model of the relationship between TYK2 (rs34536133) with reduced risk for psoriasis, psoriatic arthritis, rheumatoid arthritis, and simultaneously increased risk of COVID_19 severity.
To cite this abstract in AMA style:
Verma A, Tsao N, Thomann L, Ho Y, Carr R, crawford D, efird J, Huffman J, Hung A, Ivey K, Iyengar S, Levin M, luoh S, Lynch J, Natarajan P, Pyarajan S, Bick a, Costa L, Genovese G, Hauger R, madduri R, Pathak G, polimanti R, Voight B, Vujkovic M, Zekavat M, Zhao H, Ritchie M, Chang K, Cho K, casas J, Tsao P, Gaziano J, ODonnell C, Damrauer S, Liao K. A Phenome-Wide Association Study of Genes Associated with COVID-19 Severity Reveals Shared Genetics with Rheumatic Conditions [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/a-phenome-wide-association-study-of-genes-associated-with-covid-19-severity-reveals-shared-genetics-with-rheumatic-conditions/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-phenome-wide-association-study-of-genes-associated-with-covid-19-severity-reveals-shared-genetics-with-rheumatic-conditions/
