Session Information
Date: Monday, November 9, 2020
Title: SLE – Treatment Poster II
Session Type: Poster Session D
Session Time: 9:00AM-11:00AM
Background/Purpose: We previously reported Phase 2 SLE trial results for obexelimab, a CD19-targeted FcγRIIb engager that suppresses B-cell activation [1]. The primary endpoint, which measured loss of baseline response to steroids, was not met, but a strong trend in favor of obexelimab and statistically significant efficacy in secondary endpoints supported further analysis. Here, we describe a novel whole blood gene expression biomarker that characterized patients with improved outcomes on obexelimab therapy.
Methods: Expression of individual genes and gene pathway scores were evaluated for 68 patients who either completed the study or terminated early for loss of response using a five-fold cross validation framework. The relevance of each gene to a potential biomarker predictive model was measured by the degree to which high or low expression identified a patient subgroup (cDx+) with greater reduction in risk of flare by obexelimab than in cDX- patients.
Results: Obexelimab treatment was associated with reduction of B-cell genes and gene-sets reflective of activated B-cells and plasma cells/plasmablasts (Figure 1), as previously reported [2]. Importantly, our cross-validated analyses determined that the baseline expression levels of 2 predictive genes, CD27 and APP, identified a cDx+ group (50% of all patients) with greatly reduced risk of flare on obexelimab (cross-validated HR=0.262, full data set HR=0.166) (Figure 2). Conversely, in the placebo group, the CDx+ subjects had higher flare risk, thus cDX+ also identifies poor-prognosis patients who might particularly benefit from obexelimab. The cDx+ group demonstrated increased obexelimab effects compared to cDx- patients over a range of clinical endpoints (SRI-4: 56% cDx+ vs 14% cDx-, SRI-6: 33% vs 6.2%, LLDAS: 50% vs 6.2%, BICLA: 33% vs 12.5%) (Figure 3). CD27, the top predictive biomarker, is highly expressed by T-naïve and memory cells. Consistent with the notion that CD27 expression is important within the T-cell lineage, increased expression of other T-cell genes was also associated with reduced flare risk, including CD28 (p=0.04), TCF7(p=0.03) and FOXP3 (p=0.05). Taken together with the reduction of B cell signatures seen on treatment, this new identification of CD27 as a potential T-cell associated predictive biomarker suggests an important therapeutic role of obexelimab in suppression of B-cell and T-cell interactions.
Conclusion: A novel two-gene classifier developed from whole blood transcriptomic data by RNA-sequencing identifies a biomarker positive group of patients with a high baseline resting and stem-like T-cell (CD27+, CD28+ and TCF7+) signature. These cDx+ patients had superior response rates compared to placebo across multiple clinical endpoints, suggesting that adaptive immune responses such as B-cell antigen presentation and/or T-cell costimulation may be key components of obexelimab effects. This exploratory biomarker analysis supports further assessment of the two gene signature as a patient stratification/companion diagnostic strategy in obexelimab clinical development for SLE and other autoimmune conditions.
[1] Merrill JT, et al Arth Rheum 2018;70 Abs S10.
[2] Merrill JT, et al EULAR 2020; Abs SAT0187
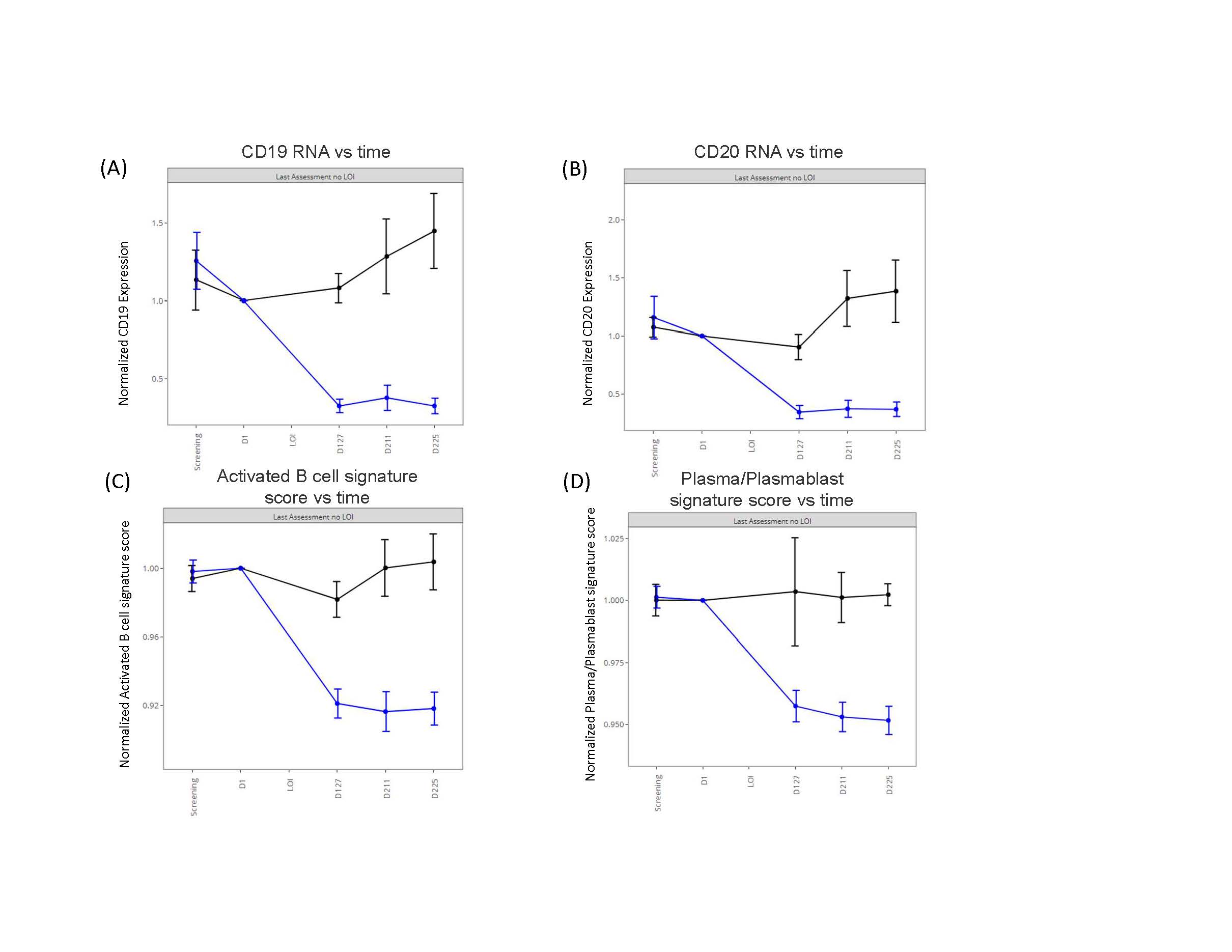 Figure 1. Pharmacodynamic effects of obexelimab (blue curve) compared with placebo (black curve) in patients with systemic lupus erythematosus (SLE) who had evaluable baseline whole blood transcriptomic (RNA-seq) data and either completed the study without a flare or experienced a flare on study. The RNA expression and signature scores are relative to baseline. Signature scores are based on immune cell signatures described in [3] and generated by the Singscore method[4] (A) B-cell marker gene CD19 gene expression over time (B) B-cell marker gene CD20 (transcript MS4A1) gene expression over time. (C) Activated B-cell signature score over time (D) Plasma/plasmablast signature score over time. [3] Arazi, A. et al. Nat. Immunol. 2019. 20;902-914. [4] Foroutan M, et al. . BMC Bioinformatics, 2018 19; 404.
Figure 1. Pharmacodynamic effects of obexelimab (blue curve) compared with placebo (black curve) in patients with systemic lupus erythematosus (SLE) who had evaluable baseline whole blood transcriptomic (RNA-seq) data and either completed the study without a flare or experienced a flare on study. The RNA expression and signature scores are relative to baseline. Signature scores are based on immune cell signatures described in [3] and generated by the Singscore method[4] (A) B-cell marker gene CD19 gene expression over time (B) B-cell marker gene CD20 (transcript MS4A1) gene expression over time. (C) Activated B-cell signature score over time (D) Plasma/plasmablast signature score over time. [3] Arazi, A. et al. Nat. Immunol. 2019. 20;902-914. [4] Foroutan M, et al. . BMC Bioinformatics, 2018 19; 404.
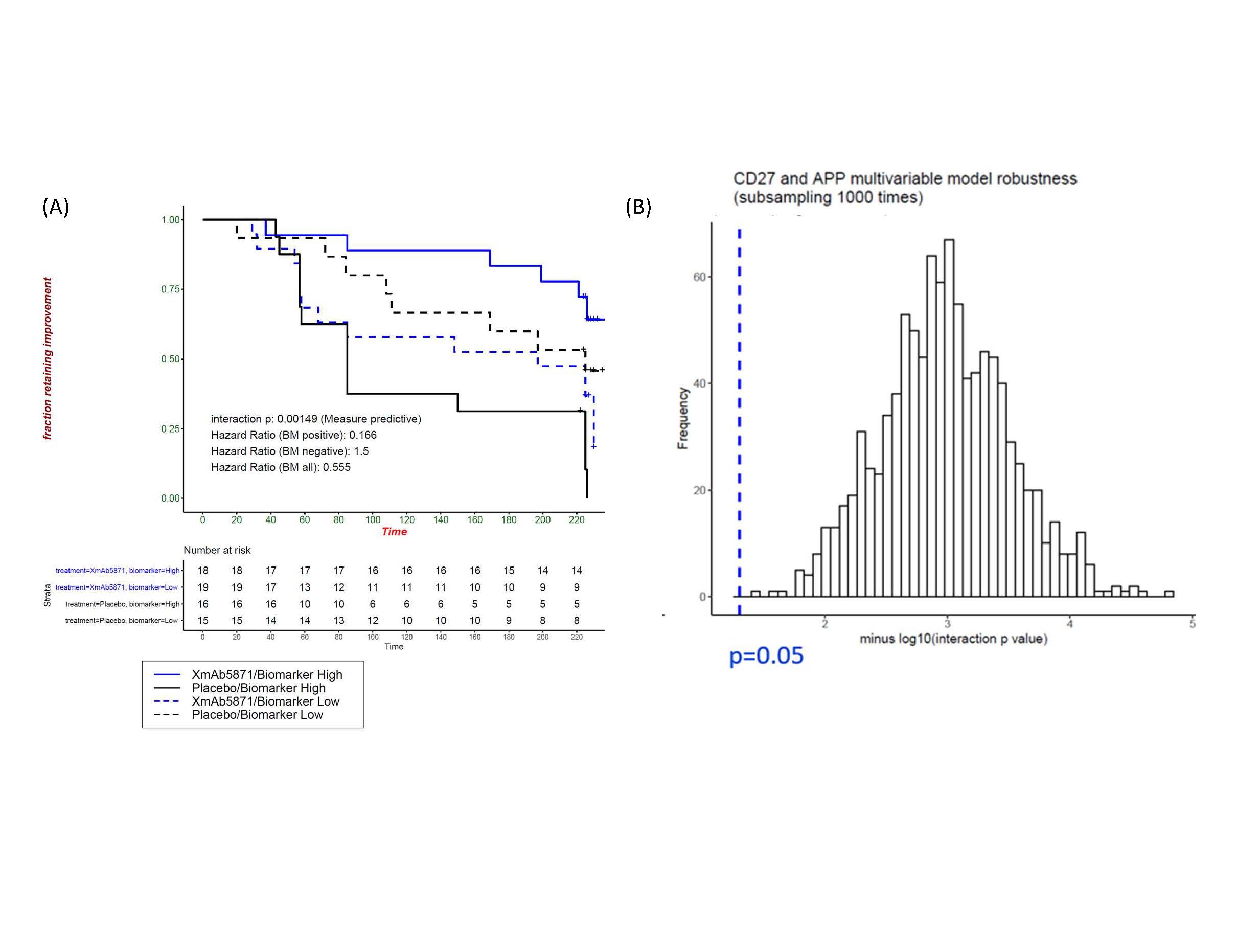 Figure 2. Two-gene predictive model (CD27 and APP). A five-fold cross validation framework was developed to ensure the generalizability of the results and to determine the number of genes needed and what fraction of patients to assign as cDx+. Classifications are based on a simple threshold for the sum of gene expression levels that have been normalized by the training sample mean and standard deviation. (A) Normalized expression levels of two genes classify patients as cDX+ or cDx-, with an estimated 50% of patients assigned to each group. The benefit from obexelimab over placebo in the cDx+ group (solid blue and solid black curves) was significantly greater than the benefit in the cDx- group (dashed blue and dashed black curves) (p=0.000149, HR cDx+ = 0.166 vs HR cDx- =1.5). (B) Robustness of the two-gene predictive model as shown by the distribution of the interaction p-value based on subsampling 80% of the datasets 1000 times.
Figure 2. Two-gene predictive model (CD27 and APP). A five-fold cross validation framework was developed to ensure the generalizability of the results and to determine the number of genes needed and what fraction of patients to assign as cDx+. Classifications are based on a simple threshold for the sum of gene expression levels that have been normalized by the training sample mean and standard deviation. (A) Normalized expression levels of two genes classify patients as cDX+ or cDx-, with an estimated 50% of patients assigned to each group. The benefit from obexelimab over placebo in the cDx+ group (solid blue and solid black curves) was significantly greater than the benefit in the cDx- group (dashed blue and dashed black curves) (p=0.000149, HR cDx+ = 0.166 vs HR cDx- =1.5). (B) Robustness of the two-gene predictive model as shown by the distribution of the interaction p-value based on subsampling 80% of the datasets 1000 times.
 Figure 3. Response rates are enriched among cDX+ (50%) patients’ four landmark endpoints at 32 weeks. (A) SRI-4 (B) SRI-6 (C) LLDAS (D) BICLA.
Figure 3. Response rates are enriched among cDX+ (50%) patients’ four landmark endpoints at 32 weeks. (A) SRI-4 (B) SRI-6 (C) LLDAS (D) BICLA.
To cite this abstract in AMA style:
Ding Y, Zack D, Burington B, Yang A, Merrill J, James J, Desjarlais J, Clynes R, Guthridge J. A Novel Biomarker Identifies Systemic Lupus Erythematous (SLE) Patients Who Benefit from Obexelimab (XmAb®5871) Treatment [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/a-novel-biomarker-identifies-systemic-lupus-erythematous-sle-patients-who-benefit-from-obexelimab-xmab5871-treatment/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-novel-biomarker-identifies-systemic-lupus-erythematous-sle-patients-who-benefit-from-obexelimab-xmab5871-treatment/
