Session Information
Date: Sunday, October 21, 2018
Title: 3S103 ACR Abstract: Genetics, Genomics & Proteomics: Precision Medicine (916–921)
Session Type: ACR Concurrent Abstract Session
Session Time: 4:30PM-6:00PM
Background/Purpose:
High-throughput gene expression profiling of skin biopsies from patients with systemic sclerosis (SSc) has identified four “intrinsic” gene expression subsets conserved across multiple cohorts and tissues. These are the inflammatory, fibroproliferative, normal-like, and limited subsets. In order to classify patients in clinical trials or for diagnostic purposes, supervised methods that can assign a single sample to a molecular subset are required. Here, we introduce a novel machine learning classifier which is a robust predictor of intrinsic subset and test it on multiple independent patient cohorts.
Methods:
Three independent gene expression cohorts were curated and merged to create a training dataset covering 297 skin biopsies from 102 SSc patients and controls to train a classifier. Supervised machine learning algorithms were rigorously trained and evaluated using repeated three-fold cross-validation. We performed external validation using three SSc cohorts (GSE66321, GSE65405, GSE58095), including a gene expression dataset generated by an independent laboratory on a different microarray platform. In total, 427 skin biopsies from 213 individuals were analyzed in the training and test cohorts. We used weighted gene co-expression network analysis and g:Profiler to identify and functionally characterize gene modules associated with the intrinsic subsets.
Results:
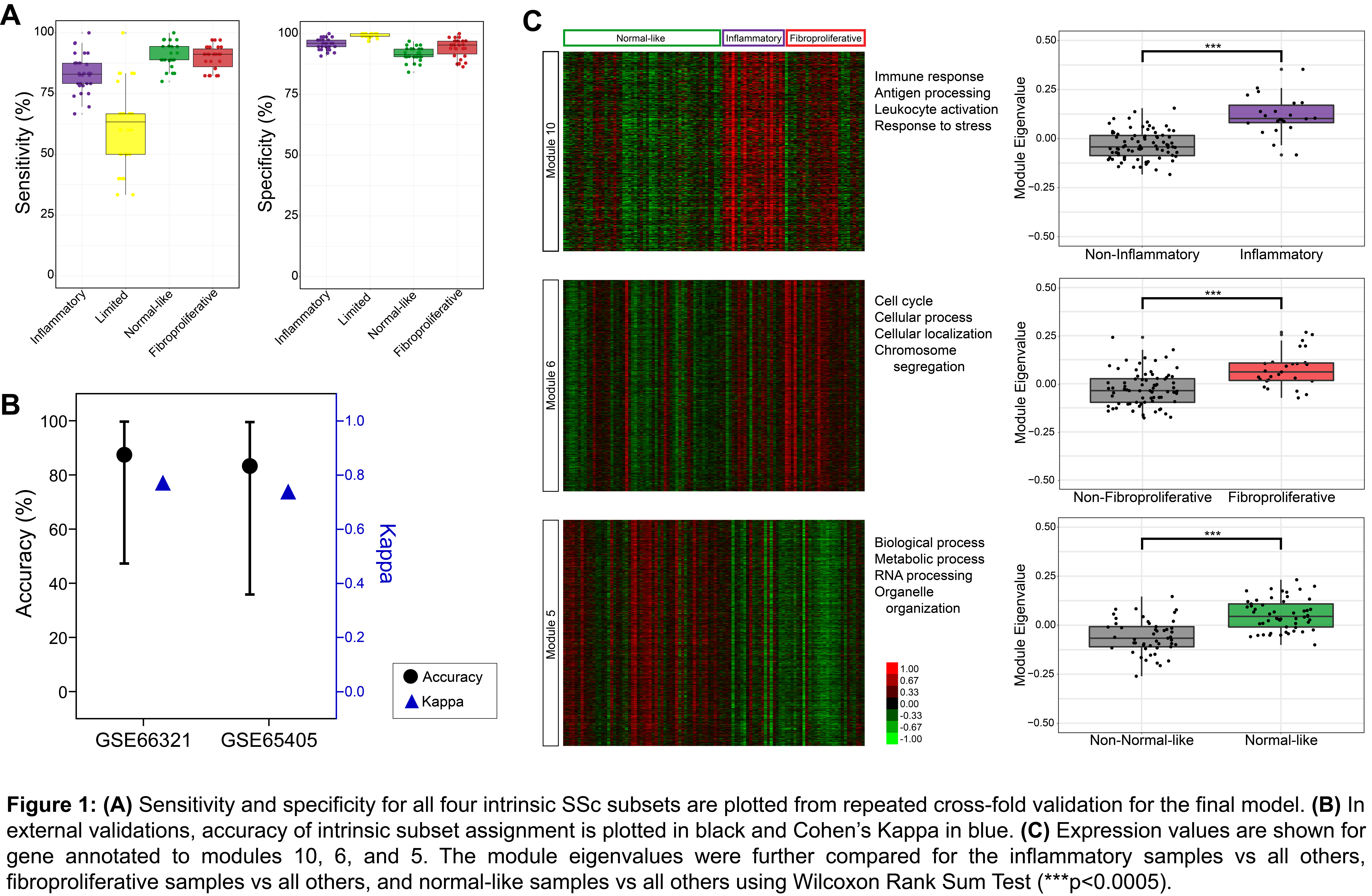
Repeated cross-fold validation identified consistent and discriminative gene expression biomarkers using multinomial elastic net, which performed with an average classification accuracy of 88.1%. All molecular subsets were classified with high sensitivity and specificity (Fig. 1A). In external validation, the classifier achieves an average accuracy of 85.4% (Fig. 1B). In a re-analysis of gene expression data from GSE58095, the classifier identified subsets of patients that represent the canonical inflammatory, fibroproliferative, and normal-like subsets (Fig. 1C). The inflammatory subset showed upregulated gene modules significantly enriched in biological processes such as inflammatory response, lymphocyte activation, and stress response. Similarly, gene modules enriched for cell cycle processes were increased in the fibroproliferative subset.
Conclusion:
We developed a highly accurate and reliable classifier for SSc molecular subsets for single samples analyzed on multiple gene expression platforms. Prior methods relied on agglomerative methods that could not be applied to single samples. These analyses show that the intrinsic gene expression subsets are a common feature of SSc found across multiple validation cohorts. Machine learning methods provide a robust and accurate mechanism for stratifying intrinsic gene expression subsets and can be used to aid clinical decision-making and interpretation for SSc patients and in clinical trials.
To cite this abstract in AMA style:
Franks J, Martyanov V, Cai G, Wang Y, Wood TA, Whitfield ML. A Machine Learning Classifier for Assigning Individual Patients with Systemic Sclerosis to Intrinsic Molecular Subsets [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/a-machine-learning-classifier-for-assigning-individual-patients-with-systemic-sclerosis-to-intrinsic-molecular-subsets/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-machine-learning-classifier-for-assigning-individual-patients-with-systemic-sclerosis-to-intrinsic-molecular-subsets/