Session Information
Session Type: Abstract Session
Session Time: 2:00PM-3:30PM
Background/Purpose: Knee osteoarthritis (KOA) is the most common form of arthritis that affects multiple knee joint tissues. The infrapatellar fat pad is the largest fat pad in the knee; however, its cell composition, including associated transcriptomic profiles, during KOA are not well described. Defining these cellular and transcriptomic profiles is crucial to defining the role of the infrapatellar fat pad in KOA pathogenesis. In this study, we subjected infrapatellar fat pad from KOA patients to high throughput single nuclei RNA sequencing (snRNAseq) and advanced bioinformatics to identify and define the cellular and transcriptomic atlas of KOA fat pad. Furthermore, we investigated transcriptomic differences in cell subsets identified in the fat pad of obese BMI patients compared to normal BMI patients with KOA.
Methods: Infrapatellar fat pad was obtained from late-stage KOA patients [KL grades III/IV; n=15; 53% females, 47% obese, BMI 30-40 and 53% non-obese, BMI 18.5-25] undergoing total knee arthroplasty and subjected to snRNAseq. Nuclei were sequenced on an Illumina NextSeq 550 using the 150bp high output sequencing kit. Cluster analysis, advanced bioinformatics and pathway analyses were employed to identify key cell types, cell subsets, cell transcriptomic profiles and biological pathways.
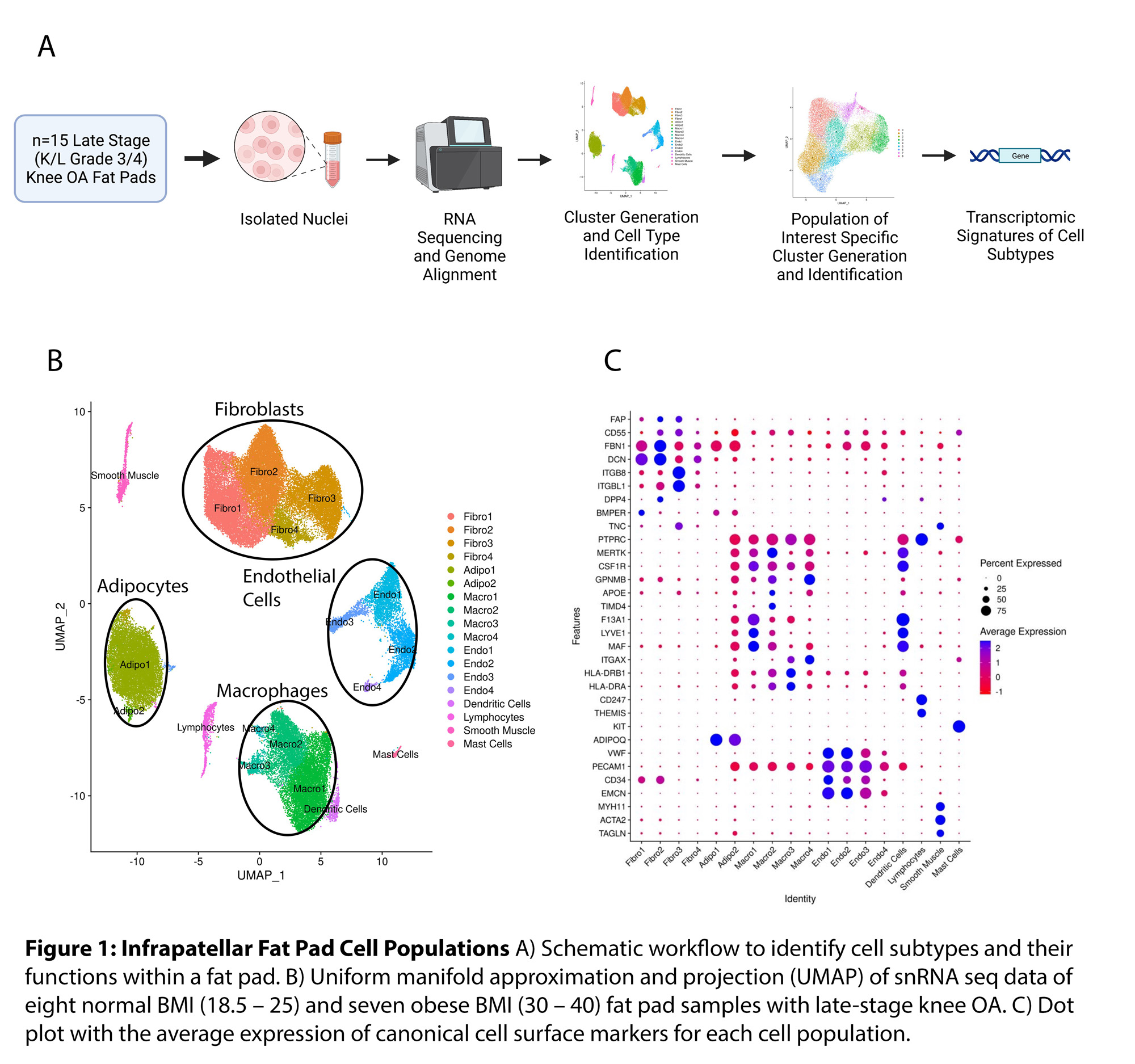
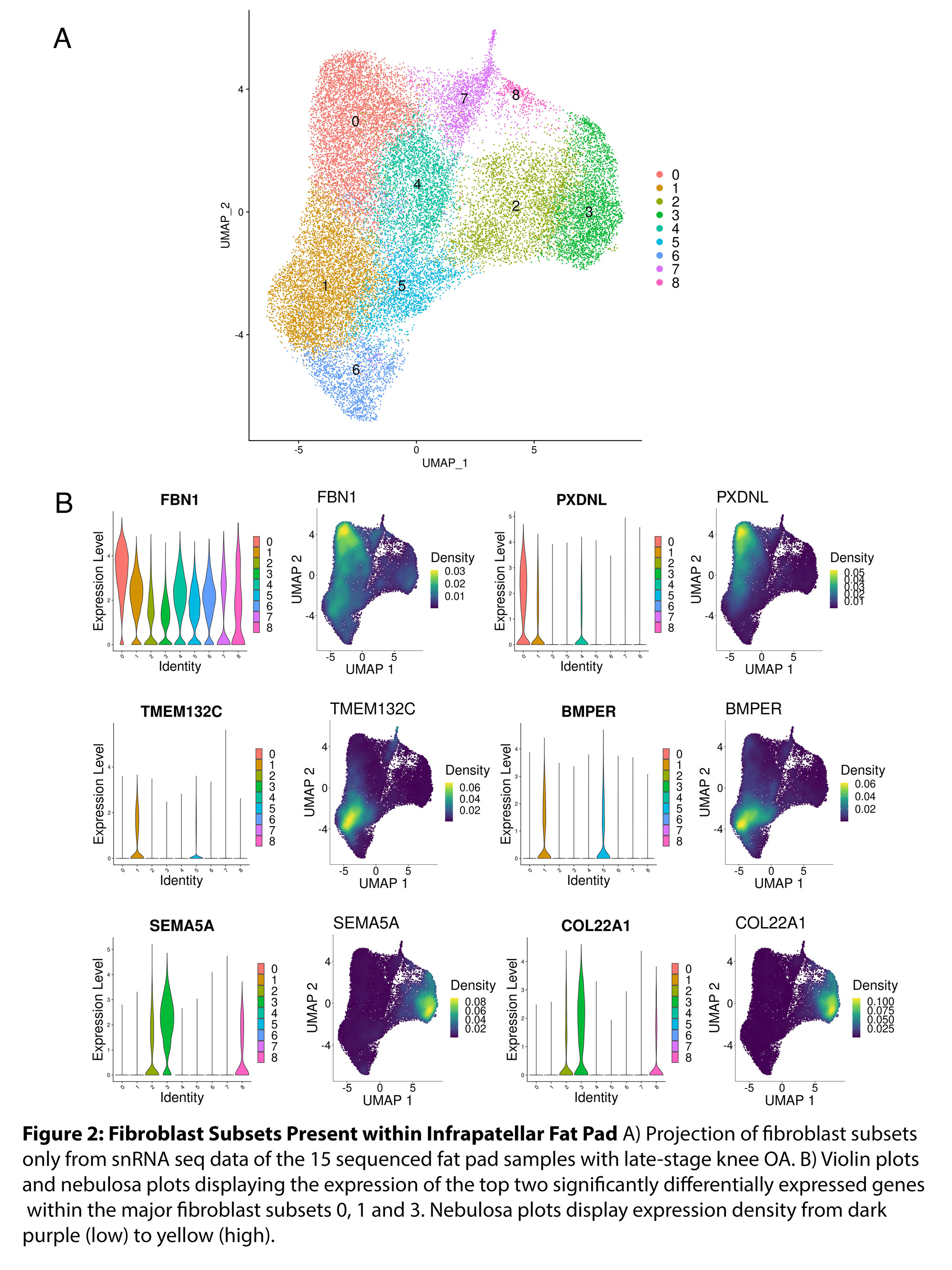
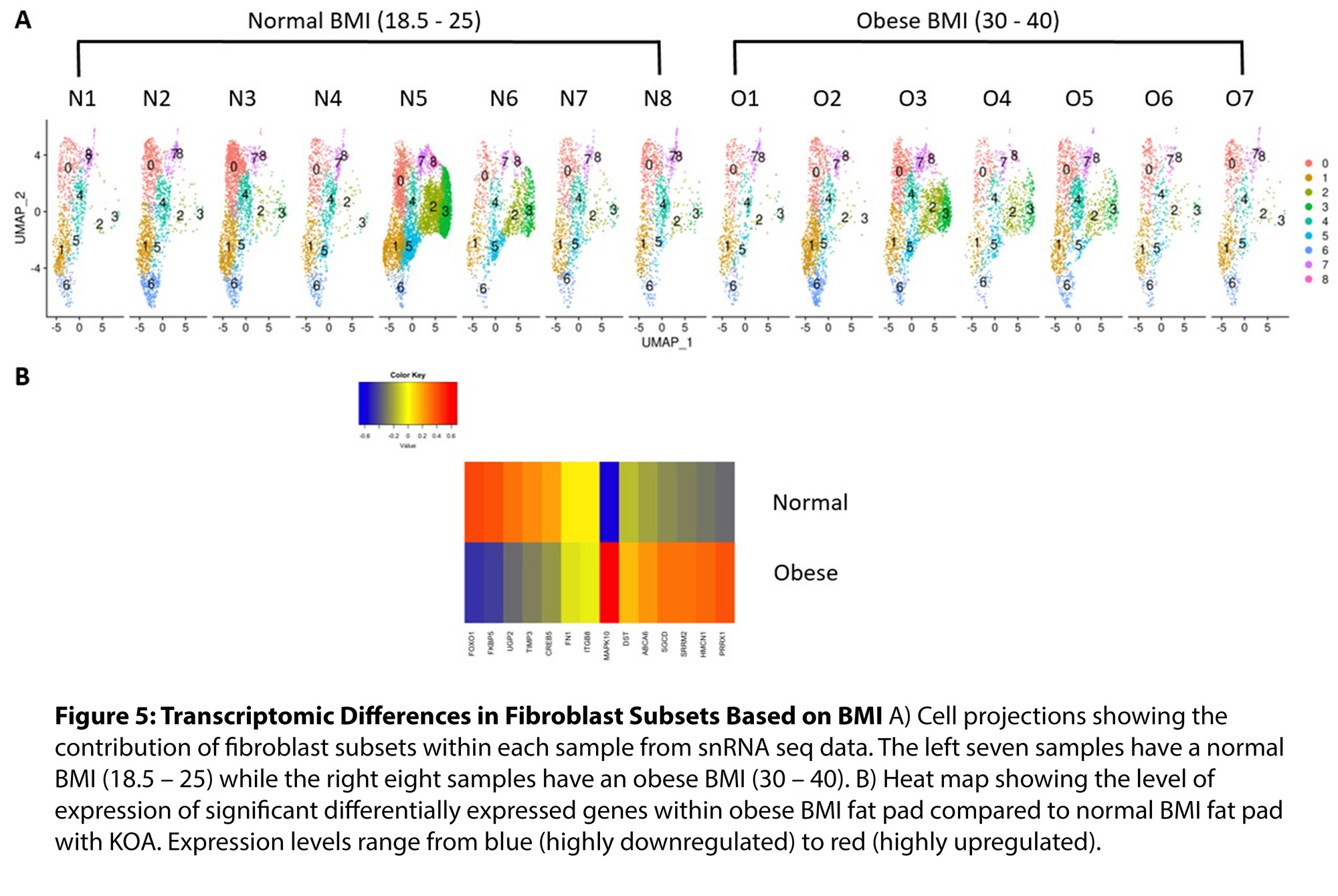
Results: Clustering analysis of snRNAseq data identified multiple cell populations based on canonical cell surface markers (Fig 1). The major cell types observed within the fat pad include fibroblasts (43.3%), macrophages (20.8%), adipocytes (18.5%), and endothelial cells (11.6%) (Fig 1). Interestingly, each major cell population identified had multiple cell subsets with unique transcriptomic profiles. Specifically, we identified nine distinct fibroblast cell subsets, four macrophage subsets, two adipocyte subsets and six endothelial subsets, each with unique transcriptomic profiles of highly expressed genes (Fig 2). We did not identify any differences in the proportions of fibroblast subsets when comparing normal BMI and obese BMI fat pads; however, we did identify a distinct transcriptomic signature with uniquely upregulated and downregulated genes in fibroblasts of the fat pad from obese BMI KOA patients compared to normal BMI KOA patients (Fig 3). We are now employing computational analyses of identified transcriptomic signatures and functional assays using primary fat pad fibroblasts of KOA patients to elucidate the mechanisms associated with the transcriptomic differences in obese BMI versus normal BMI KOA patients.
Conclusion: Using snRNAseq, we have generated a KOA fat pad transcriptomic and cell atlas. We have identified distinct cell subsets and transcriptomic profiles of fibroblasts, adipocytes, macrophages, and endothelial cells in the fat pad of patients with KOA. We also uncovered differences in transcriptomic profiles within fat pad fibroblasts of obese BMI KOA patients compared to normal BMI KOA patients. Further investigations regarding the contributions of these transcriptomic profiles and their function within the OA fat pad are currently underway.
To cite this abstract in AMA style:
Peters H, Potla P, Rockel J, Delos Santos K, Vohra S, lively s, Perry K, Perruccio A, Mahomed N, Rampersaud R, Gandhi R, Kapoor M. A Cell and Transcriptomic Atlas of the Infrapatellar Fat Pad from Patients with Knee Osteoarthritis: Identification of an Obesity-Associated Transcriptomic Signature [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/a-cell-and-transcriptomic-atlas-of-the-infrapatellar-fat-pad-from-patients-with-knee-osteoarthritis-identification-of-an-obesity-associated-transcriptomic-signature/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-cell-and-transcriptomic-atlas-of-the-infrapatellar-fat-pad-from-patients-with-knee-osteoarthritis-identification-of-an-obesity-associated-transcriptomic-signature/