Session Information
Session Type: Abstract Session
Session Time: 12:30PM-12:45PM
Background/Purpose: Familial Mediterranean Fever (FMF) is a monogenic autoinflammatory disease caused by MEFV mutations. Amyloidosis remains its most serious complication, with several risk factors reported in the literature. Traditional models, used to identify these factors, are limited by their reliance on linearity and inability to capture complex patterns in high-dimensional datasets. Machine learning ensemble models have not yet been applied to predict amyloidosis in FMF. This study aimed to develop and compare the performance of logistic regression, Random Forest and Gradient Boosting models.
Methods: From a database of 920 FMF patients diagnosed and followed at our rheumatology department between 1990–2022, records were retrieved. Patients without available documents or those followed elsewhere were excluded. Diagnoses were validated using Tel-Hashomer criteria and patients later found to have other diseases were excluded. Biopsy-confirmed amyloid fibrils were required to classify patients as having amyloidosis. Demographic, clinical, genetic, and laboratory data were recorded.Imputation with MICE method was done in R in respect to alignment of imputed dataset to original dataset. The imputed dataset loaded into Python and training-test sets (0.8:0.2) were created (random state=42). Logistic Regression, Random Forest, Gradient Boosting models were created thereafter. Recursive feature elimination was applied for logistic regression and Synthetic Minority Oversampling Technique (SMOTE) was used to oversample amyloidosis cases and prevent overfitting. Mann-Whitney and chi-squared tests were used for comparisons.
Results: Of 920 FMF patients, 615 with available data were included, 58 (9.4%) of whom had biopsy-confirmed amyloidosis. Variable comparisons between patients with and without amyloidosis are shown in Table 1. Age at symptom onset, diagnostic delay, disease duration, M694V homozygosity, frequent infections, arthritis, erysipelas-like erythema, myalgia, protracted febrile myalgia, and median CRP were significantly associated with amyloidosis (p < 0.001).Among the models, Random Forest performed best (AUC: 0.70, F1: 0.3, Precision: 0.375, Recall:0.25), followed by Gradient Boosting (AUC: 0.65, F1: 0.2, Precision: 0.17, Recall:0.25) and Logistic Regression (AUC: 0.60, F1: 0.17, Precision: 0.13, Recall:0.25). AUC curves and Shapley Additive Explanations (SHAP) values for the Random Forest model are shown in Figure 1 and Figure 2, respectively.
Conclusion: To our knowledege, this is the first study showing that ensemble machine learning models outperform logistic regression in predicting amyloidosis in FMF patients. Despite inherently “opaque” machinary and lower interpretability, these ensemble models are valuable given their superior performance. Furhtermore, increasing the Random Forest decision threshold to 0.6 raised precision to 0.75, enabling more efficient patient screening, especially in underserved areas. Advancing these models requires continued collaboration and a multidisciplinary approach. We invite the healthcare community to join in this effort to achieve meaningful progress and improved patient outcomes.
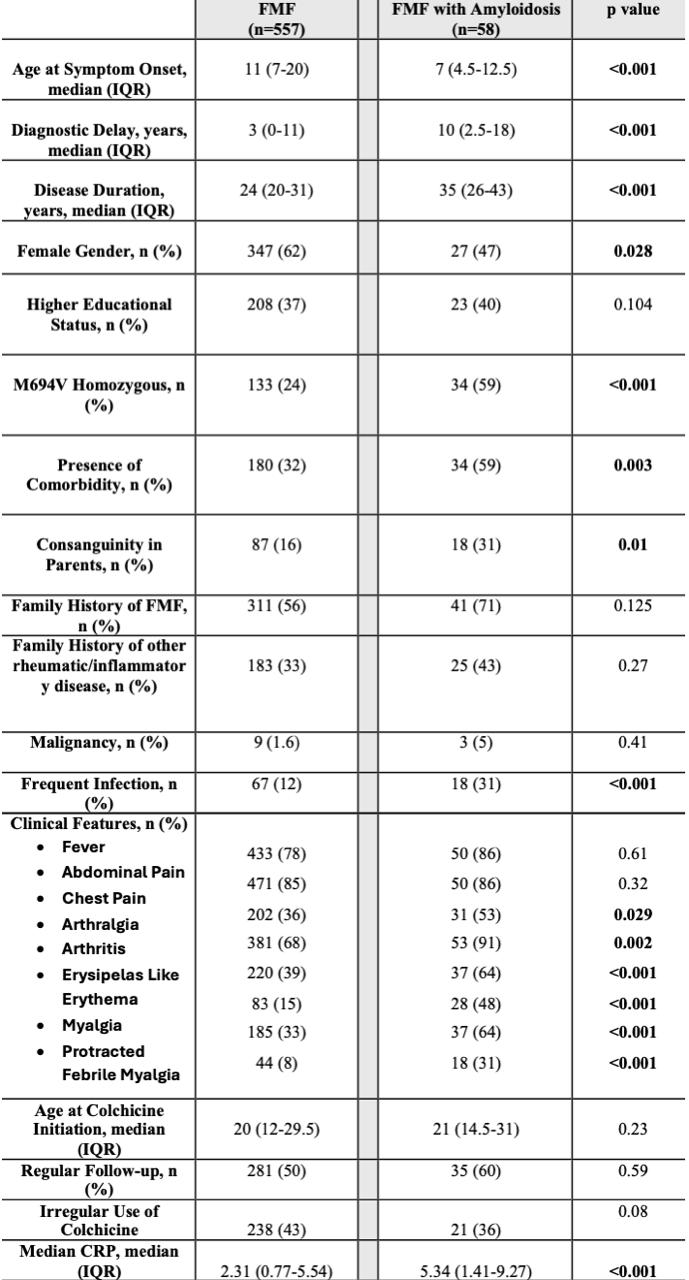 Table 1: Characteristics of the Patients with and without Amyloidosis
Table 1: Characteristics of the Patients with and without Amyloidosis
.jpg) Figure 1: AUC Curves of Models SHAP Values (Random Forest Model)
Figure 1: AUC Curves of Models SHAP Values (Random Forest Model)
.jpg) Figure 2: SHAP Values (Random Forest Model)
Figure 2: SHAP Values (Random Forest Model)
To cite this abstract in AMA style:
Aktas B, Azman E, Oner Y, Kaya k, Yuksel h, Hosgel A, Karahan D, Coban Z, parlar K, Kilinc O, Acar B, Ugurlu s. Amyloidosis Secondary to Familial Mediterranean Fever: Machine Learning Based Prediction Models [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/amyloidosis-secondary-to-familial-mediterranean-fever-machine-learning-based-prediction-models/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/amyloidosis-secondary-to-familial-mediterranean-fever-machine-learning-based-prediction-models/
