Session Information
Date: Tuesday, October 28, 2025
Title: (1830–1854) Systemic Lupus Erythematosus – Etiology and Pathogenesis Poster
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Lupus nephritis (LN) is characterized by kidney inflammation and leads to loss of kidney function. Depending on race/ethnicity, up to 60% of Systemic Lupus Erythematosus (SLE) patients develop LN early in their disease. However, the cellular pathways driving disease onset and progression remain poorly defined. Because these pathways involve cell interactions at different spatial scales, spatial mapping of kidney biopsies can provide critical insights. This study aimed to spatially profile kidney biopsies from early LN patients to identify key cellular states, interactions, and regions associated with disease progression and to construct a spatial atlas of early LN pathology.
Methods: We analyzed Formalin-Fixed Paraffin-Embedded (FFPE) kidney biopsies from patients with LN and UPCR between .250 and .499 (n = 8), non-LN SLE (no LN; n = 3), and healthy controls (n = 4). Serial sections were profiled using Xenium (5 µm; 5101 genes) and single-cell FFPE RNA seq (23 µm; whole transcriptome). Xenium transcripts were quality filtered and segmented into cells using a transcript density-based segmentation method (Baysor). Custom pipelines were used to process and integrate Xenium and scFFPE data. After QC, we obtained ~400,000 cells from Xenium and ~100,000 cells from scFFPE.
Results: Integration of scFFPE and Xenium data revealed 8 major cell lineages based on marker expression (Fig. 1A, B). LN and non-LN samples showed enrichment of lymphocytes compared to controls (Fig. 1C). Cell projections (Xenium) confirmed that profiled cells localized to expected anatomical regions (Fig. 1D). Using the public AMP SLE reference and in house pipelines, we annotated 43 cell states across the 8 cell lineages and mapped in tissue space. Transcript-based tissue segmentation with Tessera identified six regions: 3 tubular, 1 glomerular, and two interstitial (Fig. 2A, B).We next quantified cell state enrichment in each region across groups (Fig. 3). In controls, cell states localized to expected regions. In LN, macrophage subsets M5 (GPNMB high NUPR high) and M7 (SPP1 high FABP5 high) were enriched in glomerular regions, compared to interstitial enrichment in controls and non-LN SLE. Colocalization analyses revealed that M5 and M7 colocalized in the glomerular regions in LN samples, suggesting recruitment to inflamed regions during early disease.
Conclusion: We generated a comprehensive kidney spatial atlas of early LN, non-LN SLE, and healthy controls, integrating scFFPE RNA sequencing and Xenium spatial transcriptomics. In this initial study of patients with early-stage LN, we have started to characterize distinct cell states that may be involved in the development of LN in patients with SLE. We found that patients with SLE and no LN accumulated lymphocytes, suggesting that even in the absence of frank LN, the kidneys of lupus patients may be affected. We also found that resident macrophages tend to move from the interstitium to the glomerular region in patients with early LN. These macrophages may be involved in initiating glomerular injury in LN.
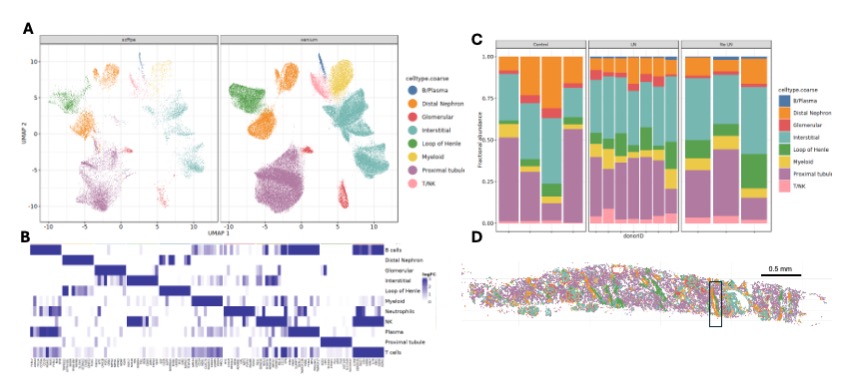 Figure1: (A) UMAP projection of cells from scFFPE RNA sequencing and Xenium datasets. Cells are colored by their major cell types. (B) Heat map of logFC of top 15 marker genes for every cell type. (C) Relative abundance of cell type populations in each sample. Relative abundance is the ratio of number of cells of each type to the total number of cells in the sample. (D) Spatial projection of the cell types in (A) onto tissue space. Insert highlights the distal tubule.
Figure1: (A) UMAP projection of cells from scFFPE RNA sequencing and Xenium datasets. Cells are colored by their major cell types. (B) Heat map of logFC of top 15 marker genes for every cell type. (C) Relative abundance of cell type populations in each sample. Relative abundance is the ratio of number of cells of each type to the total number of cells in the sample. (D) Spatial projection of the cell types in (A) onto tissue space. Insert highlights the distal tubule.
.jpg) Figure2: (A) Spatial segmentation of the tissue shown in 1D. Regions are colored by their type. Insert highlights the distal tubule in 1D, which is now detected and annotated as a DT rich region. (B) Relative abundance of cell types in each region in each sample in the dataset. Regions are annotated based on their cell composition.
Figure2: (A) Spatial segmentation of the tissue shown in 1D. Regions are colored by their type. Insert highlights the distal tubule in 1D, which is now detected and annotated as a DT rich region. (B) Relative abundance of cell types in each region in each sample in the dataset. Regions are annotated based on their cell composition.
.jpg) Figure3: (A-C): Log enrichment of cell states within each spatial region: (A) in control samples, (B) No LN samples, (C) LN samples.
Figure3: (A-C): Log enrichment of cell states within each spatial region: (A) in control samples, (B) No LN samples, (C) LN samples.
To cite this abstract in AMA style:
Madhu R, Bogle R, STEIN D, Cao J, Tsoi A, Marlin C, Eisenhaure T, Izmirly P, Masson M, Wei K, Werth V, Barnas J, Brenner M, James J, Gudjonsson J, Guthridge J, Hacohen N, Ruggles K, Raychaudhuri S, Buyon J, Petri M, Rovin B, Fava A, Korsunsky i. Integrated spatial and single-cell profiling of treatment-naïve lupus nephritis biopsies [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/integrated-spatial-and-single-cell-profiling-of-treatment-naive-lupus-nephritis-biopsies/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/integrated-spatial-and-single-cell-profiling-of-treatment-naive-lupus-nephritis-biopsies/
