Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Dengue virus (DENV) is a global public health threat that is increasing in incidence and endogenous DENV infections are expected to dramatically rise in the US. DENV has four serotypes (DENV1-4) and infection with any serotype can cause two main syndromes: 1) mild disease characterized by non-specific symptoms including fever, rash, and musculoskeletal pain, and 2) severe disease/hemorrhagic fever that can rapidly lead to multi-organ failure and death. However, the immunologic drivers of severe disease remain poorly understood. Using serum proteomics and peripheral blood mononuclear cell (PBMC) single cell RNA sequencing (scRNAseq) from pediatric patients with mild and severe dengue infection we interrogated immunologic factors associated with severe disease.
Methods: In this cross-sectional study, sera and PBMCs were collected from pediatric patients with mild dengue (DENV) (Nf12) and severe dengue/hemorrhagic fever (DHF) (Nf9) at the Robert Reid Cabral Children’s and Antonio Musa District Hospitals, Dominican Republic and from age-matched healthy controls (Nf9) at the Children’s Hospital of Philadelphia. Serum proteomics was performed using the O-link Explore HT platform. 5’ scRNAseq was performed using the 10X Genomics platform. Data analysis was performed in R.
Results: Principal component analysis of ~5000 serum proteins revealed 3 distinct clusters that separate DHF, DENV, and healthy controls (Figure 1A). DHF patients showed upregulation of multiple biomarkers consistent with Macrophage Activation Syndrome (MAS) physiology (Figure 1B). Additionally, pathway analysis performed on the significantly differentially expressed proteins (DEPs) comparing DHF to DENV demonstrated complement activation was the most significantly altered pathway (Figure 1C). Examination of the specific proteins that drive this difference showed widespread consumption of complement proteins across the complement cascade (Figure 1D). scRNAseq analysis showed increased IFNG transcription in IFNG-positive cells in DHF compared to DENV, consistent with the serum protein MAS markers (Figure 2A,B). Furthermore, the monocyte cluster is altered in DHF compared to DENV and healthy controls (Figure 2B). Re-clustering of the monocytes showed an increase in 4 monocyte subclusters in DHF (Figure 2C). Differential expression analysis to identify the top genes that define each cluster revealed that one of these clusters was characterized by early complement gene expression (C1QA, C1QB, C1QC) (Figure 2D).
Conclusion: We have identified MAS and complement dysregulation as unique features of severe dengue. Serum proteomics showed elevated IFNg and CXCL9 as well as consumption of complement proteins and loss of complement regulatory proteins that typically constrain complement activation. IFNG was also increased at the transcriptional level. Moreover, monocytes expressing early complement components are overrepresented in severe dengue patients. These findings may unveil factors that predispose to risk of developing severe dengue and support precision immunomodulation with either MAS directed or complement inhibiting therapies in severe dengue.
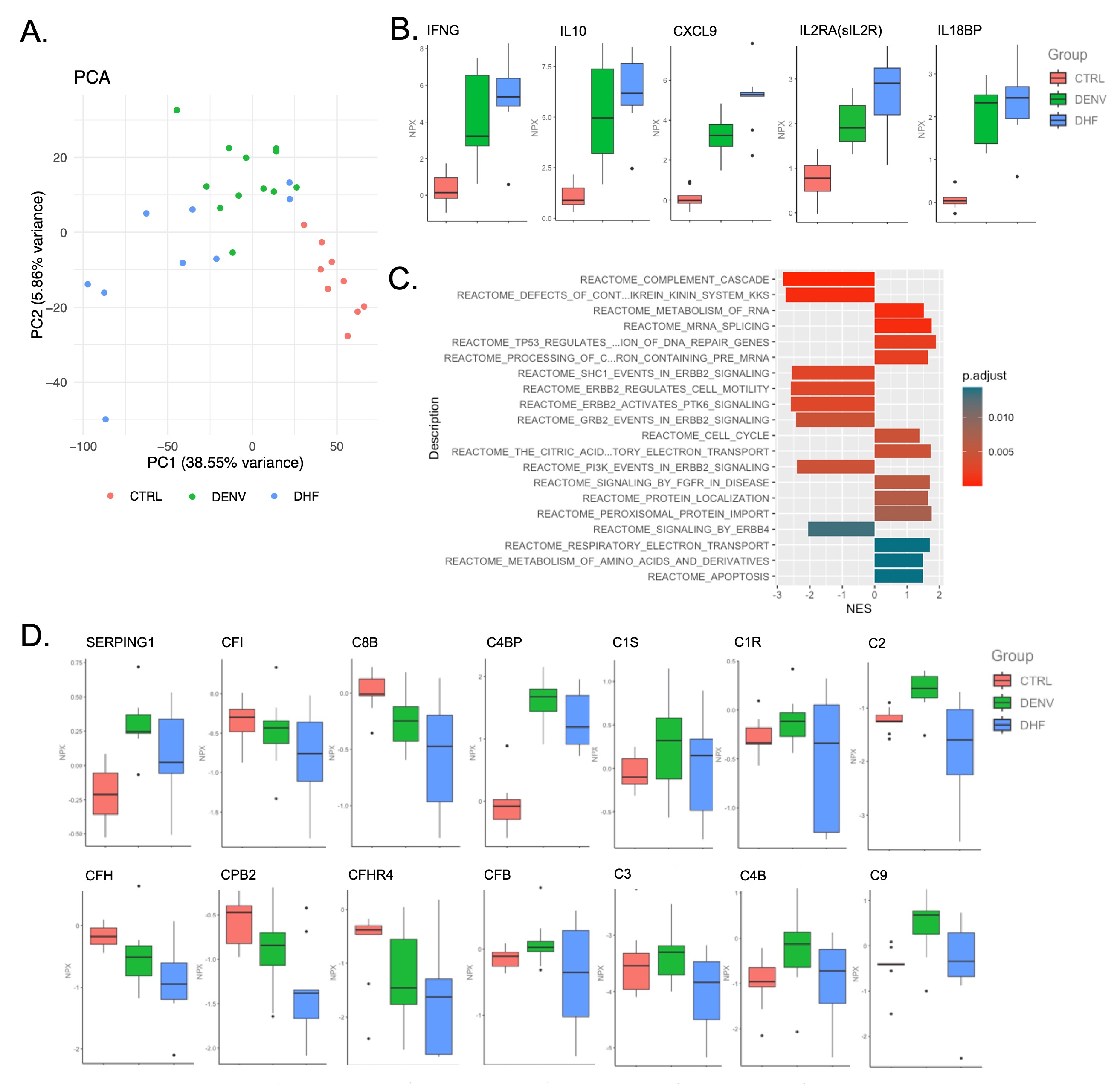 Figure 1 – Serum proteomic analysis reveals upregulation of MAS biomarkers and complement dysregulation are features of severe dengue. (A) Principal component analysis of serum proteomic data from pediatric control (CTRL, Nf9), mild dengue infection (DENV, Nf12), and severe dengue/dengue hemorrhagic fever (DHF, Nf9). (B) Box and whisker plots showing expression of the MAS associated proteins IFNG, IL10, CXCL9, IL2RA (sIL2R), IL18BP. (C) Reactome pathway analysis of significantly differentially expressed proteins comparing DHF to DENV. (D) Box and whisker plots of expression of the individual proteins that make up the Reactome Complement Cascade pathway. NPX = O-link’s Normalized Protein Expression.
Figure 1 – Serum proteomic analysis reveals upregulation of MAS biomarkers and complement dysregulation are features of severe dengue. (A) Principal component analysis of serum proteomic data from pediatric control (CTRL, Nf9), mild dengue infection (DENV, Nf12), and severe dengue/dengue hemorrhagic fever (DHF, Nf9). (B) Box and whisker plots showing expression of the MAS associated proteins IFNG, IL10, CXCL9, IL2RA (sIL2R), IL18BP. (C) Reactome pathway analysis of significantly differentially expressed proteins comparing DHF to DENV. (D) Box and whisker plots of expression of the individual proteins that make up the Reactome Complement Cascade pathway. NPX = O-link’s Normalized Protein Expression.
.jpg) Figure 2 – scRNAseq of PBMCs from patients with dengue infection shows increased IFNG expression and an altered monocyte compartment with overrepresentation of a subpopulation expressing early complement genes in severe dengue. (A) UMAP projection overlayed with IFNG transcription in mild dengue infection (DENV, Nf9) and severe dengue/dengue hemorrhagic fever (DHF, Nf6). (B) Violin plot of IFNG expression among all IFNG positive cells in DENV (Nf9) and DHF (Nf6). P value determined by unpaired T-test. (C) UMAP projection showing Azimuth l1 annotated cell types stratified by disease state; pediatric control (control, Nf5), DENV (Nf9), DHF (Nf6). (D) UMAP projection showing Seurat clusters following re-clustering of the Monocyte population, stratified by disease state; control (Nf5), DENV (Nf9), DHF (Nf6). (E) Heatmap of top differentially expressed genes that define each of the Monocyte Seurat Clusters. Clusters 1,3,4,5 are overrepresented in DHF (marked by red arrows). The red box highlights the genes that are uniquely upregulated in Cluster 1, which are composed of the genes for the C1q complement complex.
Figure 2 – scRNAseq of PBMCs from patients with dengue infection shows increased IFNG expression and an altered monocyte compartment with overrepresentation of a subpopulation expressing early complement genes in severe dengue. (A) UMAP projection overlayed with IFNG transcription in mild dengue infection (DENV, Nf9) and severe dengue/dengue hemorrhagic fever (DHF, Nf6). (B) Violin plot of IFNG expression among all IFNG positive cells in DENV (Nf9) and DHF (Nf6). P value determined by unpaired T-test. (C) UMAP projection showing Azimuth l1 annotated cell types stratified by disease state; pediatric control (control, Nf5), DENV (Nf9), DHF (Nf6). (D) UMAP projection showing Seurat clusters following re-clustering of the Monocyte population, stratified by disease state; control (Nf5), DENV (Nf9), DHF (Nf6). (E) Heatmap of top differentially expressed genes that define each of the Monocyte Seurat Clusters. Clusters 1,3,4,5 are overrepresented in DHF (marked by red arrows). The red box highlights the genes that are uniquely upregulated in Cluster 1, which are composed of the genes for the C1q complement complex.
To cite this abstract in AMA style:
McCuaig S, Gallagher M, Soto Albrecht Y, Acosta F, Turbi-Cornielle S, Soriano R, Gonzalez-Diaz E, Sanchez V, Herrera E, Cornielle A, Fernandez A, LaMonte A, Henrickson S, Vella L, Steenhoff A, Behrens E. Serum proteomic and single cell RNA sequencing analysis reveals macrophage activation syndrome physiology and widespread complement dysregulation are associated with severe dengue infection in a pediatric cohort [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/serum-proteomic-and-single-cell-rna-sequencing-analysis-reveals-macrophage-activation-syndrome-physiology-and-widespread-complement-dysregulation-are-associated-with-severe-dengue-infection-in-a-pedia/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/serum-proteomic-and-single-cell-rna-sequencing-analysis-reveals-macrophage-activation-syndrome-physiology-and-widespread-complement-dysregulation-are-associated-with-severe-dengue-infection-in-a-pedia/
