Session Information
Session Type: Abstract Session
Session Time: 1:15PM-1:30PM
Background/Purpose: Tumor necrosis factor inhibitors (TNFi) are cornerstones of autoimmune‑disease therapy, yet many patients switch agents because of loss of effectiveness, adverse events, or insurance barriers. Unstructured clinical notes contain the rationale for switching but are costly to review manually. Our objective was to determine whether large language models (LLMs) can automatically identify TNFi-to-TNFi switching and the reasons for switching in real‑world practice.
Methods: We conducted a retrospective observational study of de‑identified electronic health records (2012‑2023) from a single academic center. We defined TNFi-to-TNFi switching as a change from one TNFi to a different TNFi at consecutive encounters and include patients with a TNFi-to-TNFi switch and ≥6 months follow‑up. Using a zero‑shot prompt, GPT‑4‑turbo‑128k and eight open‑source LLMs extracted (1) TNFi stopped, (2) TNFi started, and (3) reason category between: adverse event, anti-drug antibodies, insurance/cost, lack of effectiveness, patient preference, other, or unknown. Performance was assessed on a subset of manually expert-annotated notes with micro-F1 scores.
Results: 2,112 patients met inclusion criteria (Table 1). On the manually annotated subset (n=146), GPT‑4 achieved micro‑F1 scores of 0.75 for stopped TNFi, 0.80 for started TNFi, and 0.83 for reason extraction. The best open‑source models approached GPT‑4 for medication identification: Starling‑7B‑beta (0.88 stopped, 0.79 started) and Llama‑3‑8B‑Instruct (0.82 stopped, 0.75 started). Only Llama‑3‑8B‑Instruct matched GPT‑4 for reason identification with F1=0.83. Figure 1 depicts a direct comparison between models. In the first switch, the leading reasons were lack of effectiveness (59%), adverse events (12%), and insurance/cost issues (11%). Subsequent switches showed a higher share of adverse events and patient preference and fewer effectiveness and cost‑driven changes (Figure 2).
Conclusion: GPT‑4 can extract complex TNFi switching patterns from clinical notes with high accuracy, substantially reducing the need for manual chart review. Several local models offer comparable performance. This capability enables scalable pharmacoepidemiologic studies of treatment lines and could power point‑of‑care tools that summarize medication histories for clinicians. Further work should evaluate generalizability to other biologic classes and multi‑institution datasets.
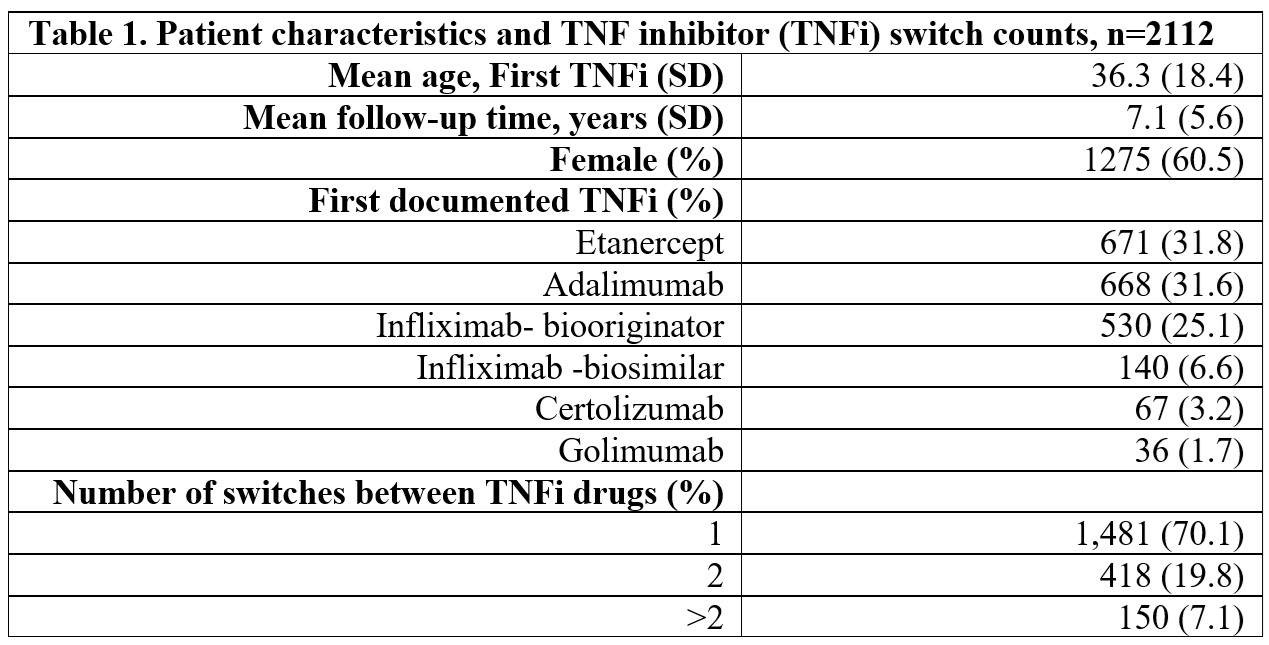 Table 1. Patient characteristics and TNF inhibitor (TNFi) switch counts, n=2112.
Table 1. Patient characteristics and TNF inhibitor (TNFi) switch counts, n=2112.
.jpg) Figure 1. Average win minus loss percentage rates of open-source large language models compared to GPT-4 in identifying started and stopped medication on the manually reviewed note subset. Positive numbers indicate the model is superior to GPT-4 (the model correctly identified a started or stopped medication where GPT-4 did not), negative numbers indicate the contrary.
Figure 1. Average win minus loss percentage rates of open-source large language models compared to GPT-4 in identifying started and stopped medication on the manually reviewed note subset. Positive numbers indicate the model is superior to GPT-4 (the model correctly identified a started or stopped medication where GPT-4 did not), negative numbers indicate the contrary.
.jpg) Figure 2. Percentage of identified reasons for switching.
Figure 2. Percentage of identified reasons for switching.
To cite this abstract in AMA style:
Miao B, Binvignat M, Garcia-Agundez A, Bravo M, Williams C, Miao C, Alaa A, Rudrapatna V, Schmajuk g, Yazdany J. Extracting TNF Inhibitor Switching Reasons and Trajectories From Real-World Data Using Large Language Models [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/extracting-tnf-inhibitor-switching-reasons-and-trajectories-from-real-world-data-using-large-language-models/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/extracting-tnf-inhibitor-switching-reasons-and-trajectories-from-real-world-data-using-large-language-models/
