Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Rheumatoid arthritis (RA) is a chronic autoimmune disease in which biologic and targeted synthetic DMARDs (bDMARDs, tsDMARDs) have substantially improved clinical outcomes. While randomized controlled trials (RCTs) support their clinical use, RCTs have limitations in cost, generalizability, and long-term risk assessment. Mendelian randomization (MR), using genetic variants as proxies for drug target modulation, provides an alternative approach for inferring causality. This study aimed to assess whether genetically proxied modulation of known therapeutic targets is causally associated with RA risk, thereby validating drug efficacy from a genetic perspective.
Methods: We applied a two-sample drug-target MR framework using cis-eQTLs from the eQTLGen Consortium for 18 gene targets corresponding to approved RA therapies. Outcome datasets included three large RA GWASs: a pan-European cohort (ieu-a-834), and two Finnish FinnGen cohorts stratified by seropositivity. Three MR methods (inverse variance weighted, MR-Egger, weighted median) were applied. Sensitivity analyses included MR-PRESSO, Steiger directionality, heterogeneity testing, and Bayesian colocalization. Results were interpreted based on convergence across methods, consistency in directionality, and robustness to pleiotropy.
Results: Across three independent datasets, TNF, FCGR2B, IL1R1, and JAK1 consistently demonstrated statistically significant and directionally concordant associations with RA risk, affirming their causal relevance. TNF and JAK1 were negatively associated with RA, consistent with their known therapeutic effects. FCGR2B and IL1R1 also showed strong associations, supporting their roles as immune regulators. Colocalization analysis identified a shared causal variant for CTLA4 in the pan-European cohort (PP.H4 = 1.0), while TNF and LTA showed distinct variants. Other genes such as MS4A1 (CD20), IL6R, and CTLA4 demonstrated inconsistent or dataset-specific effects, indicating population-dependent signals or context-specific relevance. A multitarget MR of abatacept-related genes (CD80, CD86, CTLA4) also showed a significant protective effect. Discrepancy analysis revealed that beta estimates and standard errors varied by cohort, underscoring the importance of dataset selection.
Conclusion: This study is the first to apply a systematic drug-target MR framework to validate approved RA therapies. MR reliably recapitulated the effects of known therapeutic targets such as TNF, JAK1, and FCGR2B. Integration with colocalization analysis enhanced causal interpretation, particularly for CTLA4. However, inconsistent findings for MS4A1 and IL1R1 across cohorts highlight the need for population-specific and tissue-specific validation. Our findings demonstrate the utility of MR in therapeutic target evaluation and support its use alongside RCT data in future drug development and repurposing strategies.
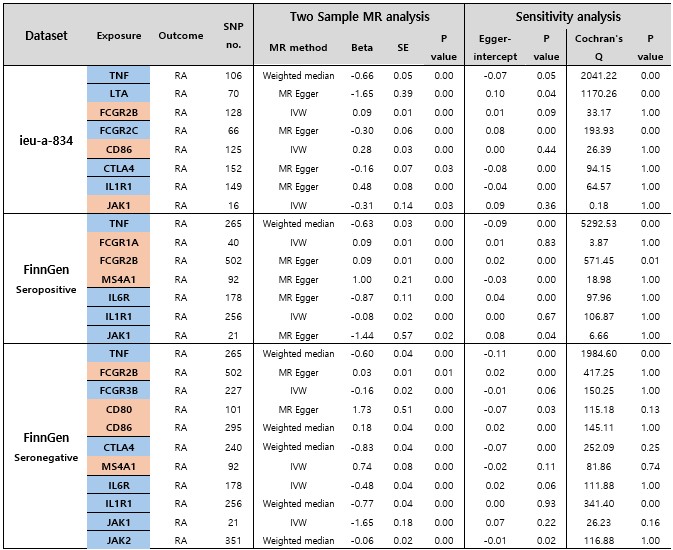 Table 1. Summary of Mendelian Randomization estimates and results from sensitivity analyses.
Table 1. Summary of Mendelian Randomization estimates and results from sensitivity analyses.
.jpg) Figure 1. Forest plots of Mendelian randomization estimates for 18 candidate genes across three rheumatoid arthritis GWAS datasets
Figure 1. Forest plots of Mendelian randomization estimates for 18 candidate genes across three rheumatoid arthritis GWAS datasets
.jpg) Figure 2. Venn diagram showing genes significantly associated with RA in each dataset
Figure 2. Venn diagram showing genes significantly associated with RA in each dataset
To cite this abstract in AMA style:
Kwon M, Joung C, Song Y. Evaluating Causal Effects of Biologic and Targeted Synthetic Disease-modifying Anti-rheumatic Drugs Targets in Rheumatoid Arthritis Using Mendelian Randomisation [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/evaluating-causal-effects-of-biologic-and-targeted-synthetic-disease-modifying-anti-rheumatic-drugs-targets-in-rheumatoid-arthritis-using-mendelian-randomisation/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/evaluating-causal-effects-of-biologic-and-targeted-synthetic-disease-modifying-anti-rheumatic-drugs-targets-in-rheumatoid-arthritis-using-mendelian-randomisation/
