Session Information
Date: Monday, October 27, 2025
Title: (1306–1346) Rheumatoid Arthritis – Diagnosis, Manifestations, and Outcomes Poster II
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: A treat-to-target approach in rheumatoid arthritis (RA) requires intense monitoring of RA disease activity with measures such as the clinical disease activity index (CDAI). Outside of clinical studies, CDAI may be sparsely recorded by clinicians in real world settings due to their local electronic health record (EHR) systems lacking systematic integration of structured entry forms for CDAI (e.g., homunculus) and variability in documentation practice among clinicians. We hypothesize that most clinicians document CDAI in unstructured free text. Two previous publications developed successful natural language processing (NLP) pipelines extracting CDAI, but the practices were limited to community settings and the Veterans Administration. We explored extracting CDAIs from a large, tertiary academic center EHR with a heterogeneous RA patient population utilizing novel NLP techniques.
Methods: The New York Presbyterian/Columbia University Medical Center Clinical Data Warehouse (NYP/CUMC CDW) contains over 4.5 million patients’ data stored in structured and unstructured formats. From a pre-selected group of RA patients in the CDW, a random sample (20%) of free text notes recorded in rheumatology outpatient practices across one EHR system (EPIC) between 2020-2024 were produced. A list of CDAI, its key components, as well as plausible variations in phrasing was generated with expert curation (EP) (Table 1). These keywords, along with serostatus (seropositivity) were extracted using an automated information extraction pipeline, engineered through large language model (LLM) prompts input through Chat GPT-4 Education, a HIPAA compliant, CUMC approved resource (Figure 1).
Results: A total of 1,983 unique RA patients (one note per patient closest to July 1, 2024) were analyzed; 768 (38.7%) were seropositive, 562 (28.3%) were seronegative, and 649 (32.7%) did not have serostatus recorded. The term “CDAI” and its variations were extracted from 173 (8.72%) patients with a median value of 7 (range 0-43). Of the 173 with CDAI recorded, 59 (34.1%) were in remission, 51 (29.4%) had low activity, 31 (17.9%) had moderate activity, and 31 (17.9%) had high activity. Of those with CDAI recorded, 137 (79.1%) were seropositive and 35 (20.2%) were seronegative (Table 2). An expert (EP) performed a chart review of a random 20% sample of the extracted CDAI scores, resulting in a precision of 0.97, recall of 0.97, and F1 score of 0.97.
Conclusion: We demonstrated feasibility and accuracy of a Chat GPT-4 supported NLP/LLM pipeline to extract CDAI scores from a large, academic EHR. At our institution, CDAI appears to be sparsely documented (< 10%) in the sampled portion of notes. Our next steps include: 1) Refining the RA cohort by chart validating those without serostatus (i.e., confirming if they are actual RA cases); 2) Integrating analysis of historical EHRs used prior to EPIC (before 2020) to perform longitudinal extraction of CDAI scores; and 3) Exploring portability of this pipeline to other academic institutions, with the goal of external validation.
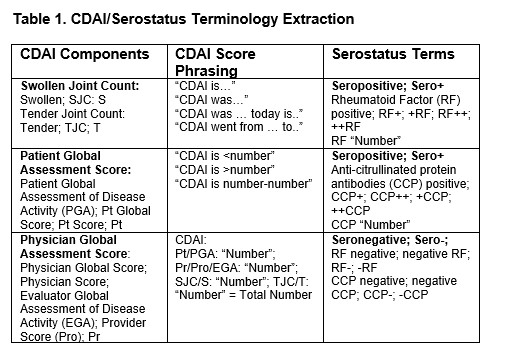 CDAI/Serostatus Terminology Extraction
CDAI/Serostatus Terminology Extraction
To cite this abstract in AMA style:
Park E, Kamdar I, Weisberg R, Nguyen A, Bathon J, Giles J, Weng C, Bernstein E. Naturalized Language Processing Based Extraction of Rheumatoid Arthritis Disease Activity Measures from the Electronic Health Record [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/naturalized-language-processing-based-extraction-of-rheumatoid-arthritis-disease-activity-measures-from-the-electronic-health-record/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/naturalized-language-processing-based-extraction-of-rheumatoid-arthritis-disease-activity-measures-from-the-electronic-health-record/

.jpg)
.jpg)