Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: ANCA-associated vasculitis frequently involves the kidneys causing glomerulonephritis (AAGN). Despite advances in treatment, many patients develop end-stage kidney disease. An improved understanding of the immune drivers of AAGN has the potential to improve treatment outcomes. To achieve that goal, we utilized a combined transcriptomics/compartmental proteomics approach to identify the major immune pathways driving AAGN.
Methods: Formalin-fixed paraffin embedded kidney biopsies of 23 patients with AAGN and 5 healthy controls (pre-implantation kidney donor biopsy) were used. For transcriptomics, RNA was extracted and bulk RNAseq was performed on all 23 AAGN biopsies and controls. Differential expression analysis was performed using the DESeq framework. For proteomics, laser capture microdissection was utilized to collect the glomerular and tubulointerstitial compartments separately using 10 AAGN biopsies and all controls. Peptides were extracted and submitted for mass-spectrometric analysis. After normalization, differential expression analysis was done using T-tests. The Benjamini-Hochberg method was used for multiple hypothesis testing correction in all experiments. Differentially abundant mRNA and peptide spectral counts were used to identify enriched immune pathways using Ingenuity pathway analysis.
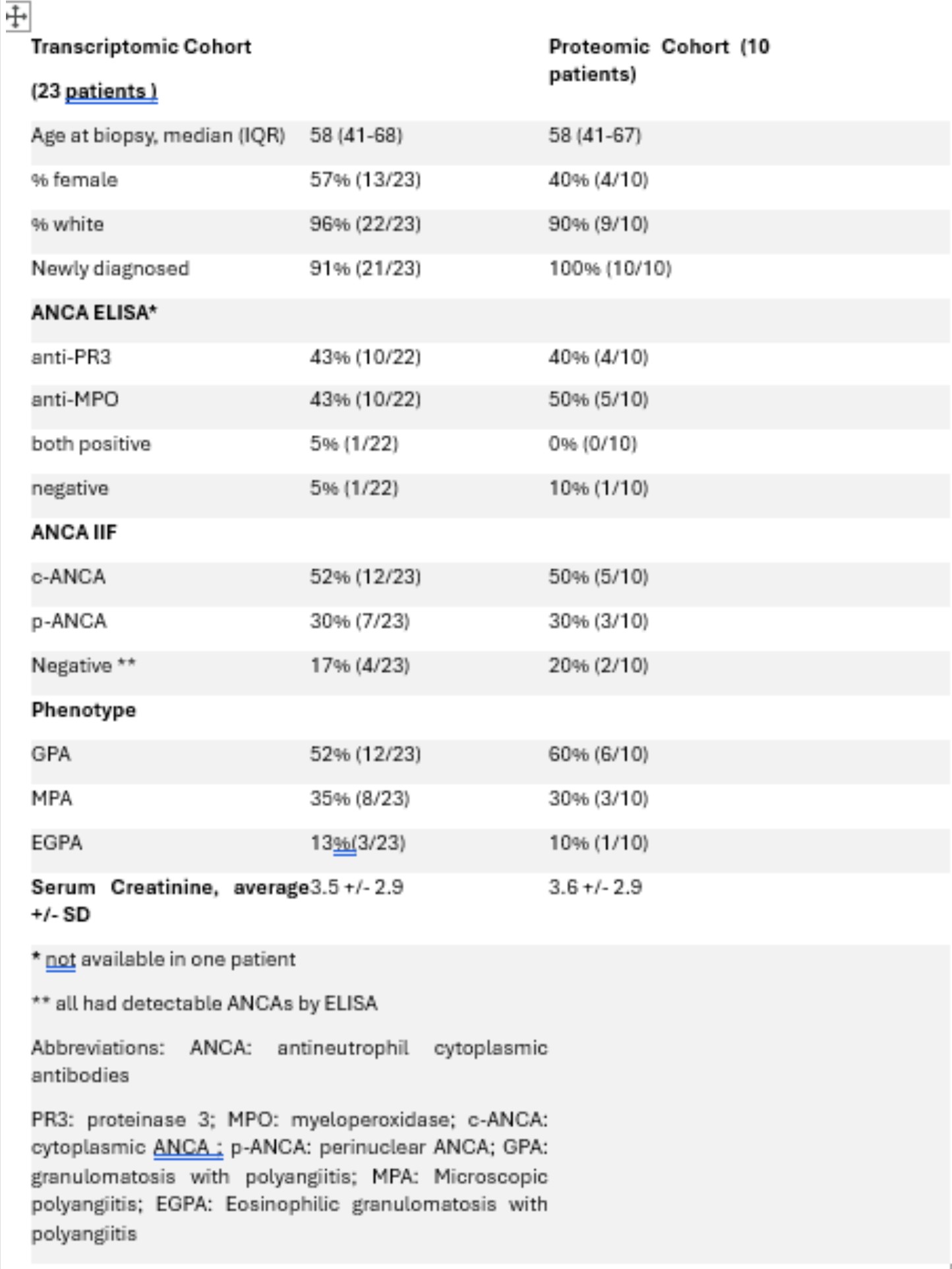
Results: Patient and disease characteristics of the full (transcriptomic) and proteomics cohort are described in table 1. The top enriched pathways in each of the 3 experiments (whole tissue transcriptomics, glomerular proteomics, tubulointerstitial proteomics) are depicted in Figure 1a-c. Comparison analysis using data from the 3 experiments demonstrated many shared enriched and upregulated pathways including complement, neutrophil degranulation, and FC gamma receptor signaling. PD-1/PDL1 and IL-10 signaling were downregulated across the 3 tissues studied. Upstream analysis demonstrated upregulation of TNF, INF-gamma, INF-alpha, and IL-6.
Conclusion: Analysis of biopsies from patients with AAGN demonstrated largely similar upregulated/downregulated immune pathways and upstream regulators when comparing transcriptomic and compartmental proteomics approaches. These may help inform patient stratification and therapy development.
.jpg) Top 20 upregulated pathways of immunologic relevance in a) transcriptomic data b) glomerular proteomic data and c) tubulointerstitial proteomic data
Top 20 upregulated pathways of immunologic relevance in a) transcriptomic data b) glomerular proteomic data and c) tubulointerstitial proteomic data
.jpg) Heatmap of identified canonical pathways and upstream regulators across transcriptomic and proteomic datasets
Heatmap of identified canonical pathways and upstream regulators across transcriptomic and proteomic datasets
To cite this abstract in AMA style:
Stojkic I, Arazi A, Song H, Yan P, Puchulu-Campanella E, Wang H, Fussner L, Rovin B, Ardoin S, Almaani S. A Combined Transcriptomics and Proteomics Approach to Identify Immune Signatures in ANCA-Associated Glomerulonephritis [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/a-combined-transcriptomics-and-proteomics-approach-to-identify-immune-signatures-in-anca-associated-glomerulonephritis/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-combined-transcriptomics-and-proteomics-approach-to-identify-immune-signatures-in-anca-associated-glomerulonephritis/