Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Severe COVID-19 infection resulting in hospitalization shares features with frequently fatal cytokine storm syndromes (CSS), such as hemophagocytic lymphohistiocytosis (HLH) and macrophage activation syndrome (MAS). Repeatedly, 30-40% of patients in published secondary (beyond infancy) HLH cohorts possess rare heterozygous mutations in established familial HLH (fHLH) genes. These mutations are thought to contribute to CSS via hypomorphic or dominant-negative effects on lymphocyte cytolytic activity. Diminished lymphocyte cytolytic activity results in prolonged lymphocyte with antigen presenting cell engagement yielding increased pro-inflammatory cytokine production contributing to multi-organ system failure.
Methods: Various published criteria were studied to define CSS among patients (n=32) enrolled in a randomized, double-blinded, placebo-controlled COVID-19 clinical trial exploring interleukin-1 blockade treatment for severe COVID-19. Whole genome sequencing (WGS) was undertaken to explore immunologic gene mutation associations among the first 20 patients enrolled in the trial for whom samples for WGS were available. Foamy viral transduction of human NK-92 natural killer (NK) cells with wild-type (control) DOCK8 cDNA or patient-derived DOCK8 missense mutations was used to explore NK cell degranulation (CD107a expression) and cytolytic activity against K562 target cells by flow cytometry.
Results: None of the patients met HLH-04 or HScore criteria, but the ferritin to erythrocyte sedimentation rate (ferritin:ESR) ratio and the COVID-19 cytokine storm syndrome score (CSS score) identified 84% and 81% of patients, respectively (Table 1). Rare mutations in fHLH genes were identified by WGS in 6 patients (30%), and 4 patients (20%) possessed rare mutations in DOCK8 (a novel CSS gene) (Table 2). The 3 patient-derived DOCK8 missense mutations introduced into NK-92 cells each diminished NK cell cytolytic function and degranulation (Figure). In addition, variants in autoinflammatory and primary immunodeficiency genes were identified in 15% and 60% of patients, respectively (Table 2).
Conclusion: This severe COVID-19 cohort, like others, shares CSS features but is best identified by ferritin:ESR ratio. Rare heterozygous CSS gene (fHLH genes and DOCK8) mutations were frequently (45%) identified in this severe COVID-19 cohort, and the DOCK8 missense mutations may have contributed to CSS in severe COVID-19 patients via diminished lymphocyte cytolytic activity.
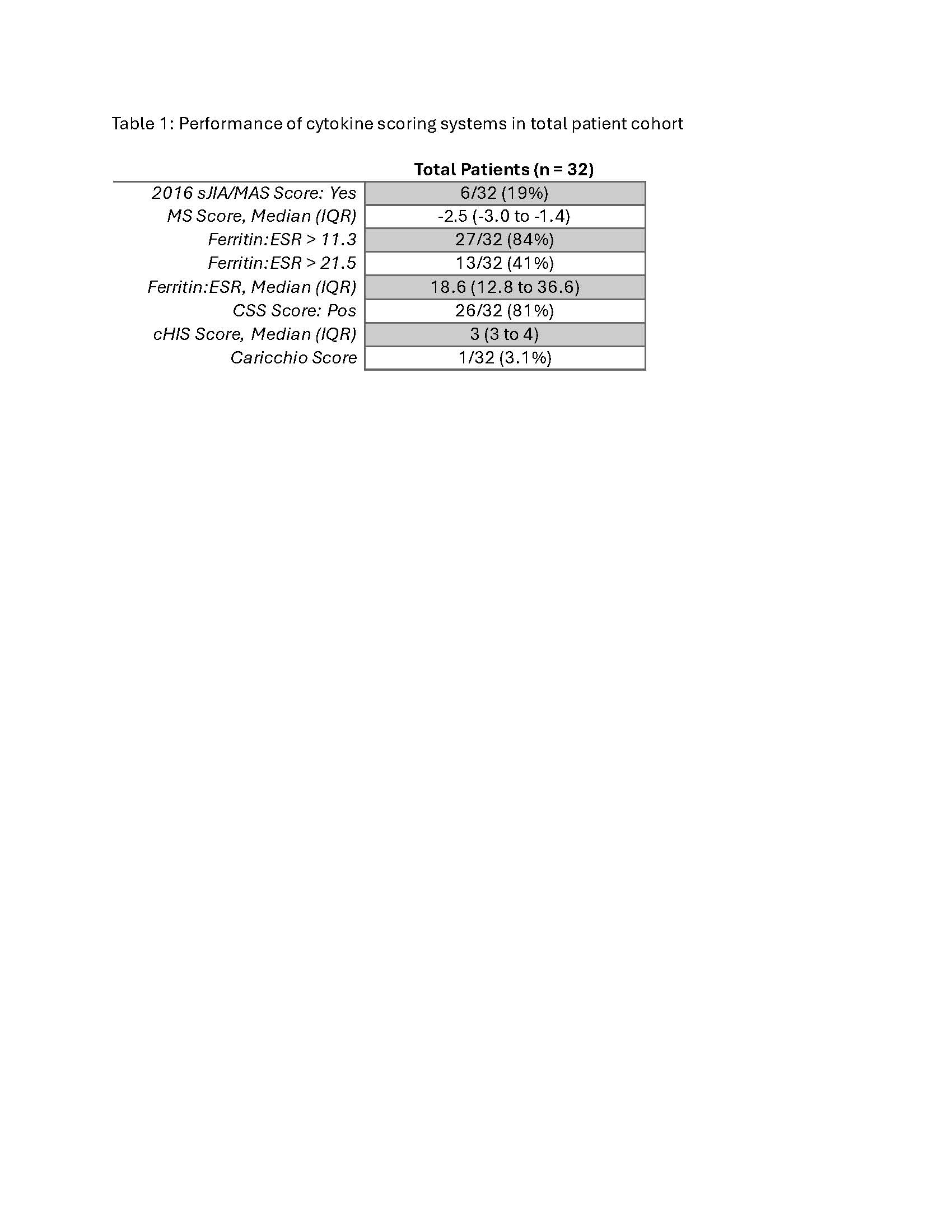 Performance of cytokine scoring systems in total patient cohort.
Performance of cytokine scoring systems in total patient cohort.
Abbreviations: sJIA – systemic juvenile idiopathic arthritis; MAS – macrophage activation syndrome; MS – MAS/sJIA; IQR – interquartile range; CSS – cytokine storm syndrome score; cHIS – COVID-19-associated hyperinflammatory syndrome
.jpg) Immunologic rare gene variants identified by whole genome sequencing of COVID-19 clinical trial participants.
Immunologic rare gene variants identified by whole genome sequencing of COVID-19 clinical trial participants.
Abbreviations: fHLH – familial hemophagocytic lymphohistiocytosis; NK – natural killer
.jpg) NK cell lytic function of DOCK8 missense variants. A. Flow cytometry plots of CD107a expression on NK-92 cells transfected with WT cDNA or patient-derived DOCK8 variants after co-incubation with K562 target cells. B. Summary plots of CD107a expression from 4 experiments. C. Flow cytometry plots of K562 target cell lysis (death) after incubation with WT DOCK8 or patient-derived variant-expressing NK-92 cells. D. Summary plots NK-92 cell cytotoxicity from 4 experiments.
NK cell lytic function of DOCK8 missense variants. A. Flow cytometry plots of CD107a expression on NK-92 cells transfected with WT cDNA or patient-derived DOCK8 variants after co-incubation with K562 target cells. B. Summary plots of CD107a expression from 4 experiments. C. Flow cytometry plots of K562 target cell lysis (death) after incubation with WT DOCK8 or patient-derived variant-expressing NK-92 cells. D. Summary plots NK-92 cell cytotoxicity from 4 experiments.
To cite this abstract in AMA style:
Cron r, Kamath A, Zhang M, Abhser D, Jackson L, Chatham W. Hemophagocytic Lymphohistiocytosis Gene Variants in Severe COVID-19 Cytokine Storm Syndrome [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/hemophagocytic-lymphohistiocytosis-gene-variants-in-severe-covid-19-cytokine-storm-syndrome/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/hemophagocytic-lymphohistiocytosis-gene-variants-in-severe-covid-19-cytokine-storm-syndrome/
