Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Childhood-onset Systemic Lupus Erythematosus (cSLE) is a clinically heterogeneous autoimmune disease. We hypothesized that similarity network fusion (SNF) a data driven method would identify relatively clinically homogeneous subgroups of patients that may represent SLE endotypes with distinct genetics.
Methods: We included patients diagnosed with cSLE between January 1992-October 2023. All patients met 2019 ACR-EULAR classification criteria and were genotyped on an Illumina multiethnic array. Ungenotyped SNPs were imputed with TopMed as a referent. We extracted SLE manifestations, date of each manifestation onset and demographic characteristics from a dedicated Lupus database. Ancestry was genetically inferred using principal components and ADMIXTURE with 1000 Genomes as a referent. We used time to each SLE manifestation onset from SLE diagnosis to identify patient subgroups with SNF. We conducted Kaplan Meir analyses and ran Cox proportional-hazard models on the time from SLE diagnosis to onset of specific manifestations between SNF clusters. We tested cluster differences in demographic and manifestation prevalences using χ2 or Fisher’s exact test, and time to each SLE manifestation onset with log rank tests. We calculated additive, allelic weighted SLE non-HLA and HLA polygenic risk scores (PRS) using alleles from a transancestral GWAS of SLE. Models adjusted for sex and ancestry determined the relationship between PRS and cluster membership.
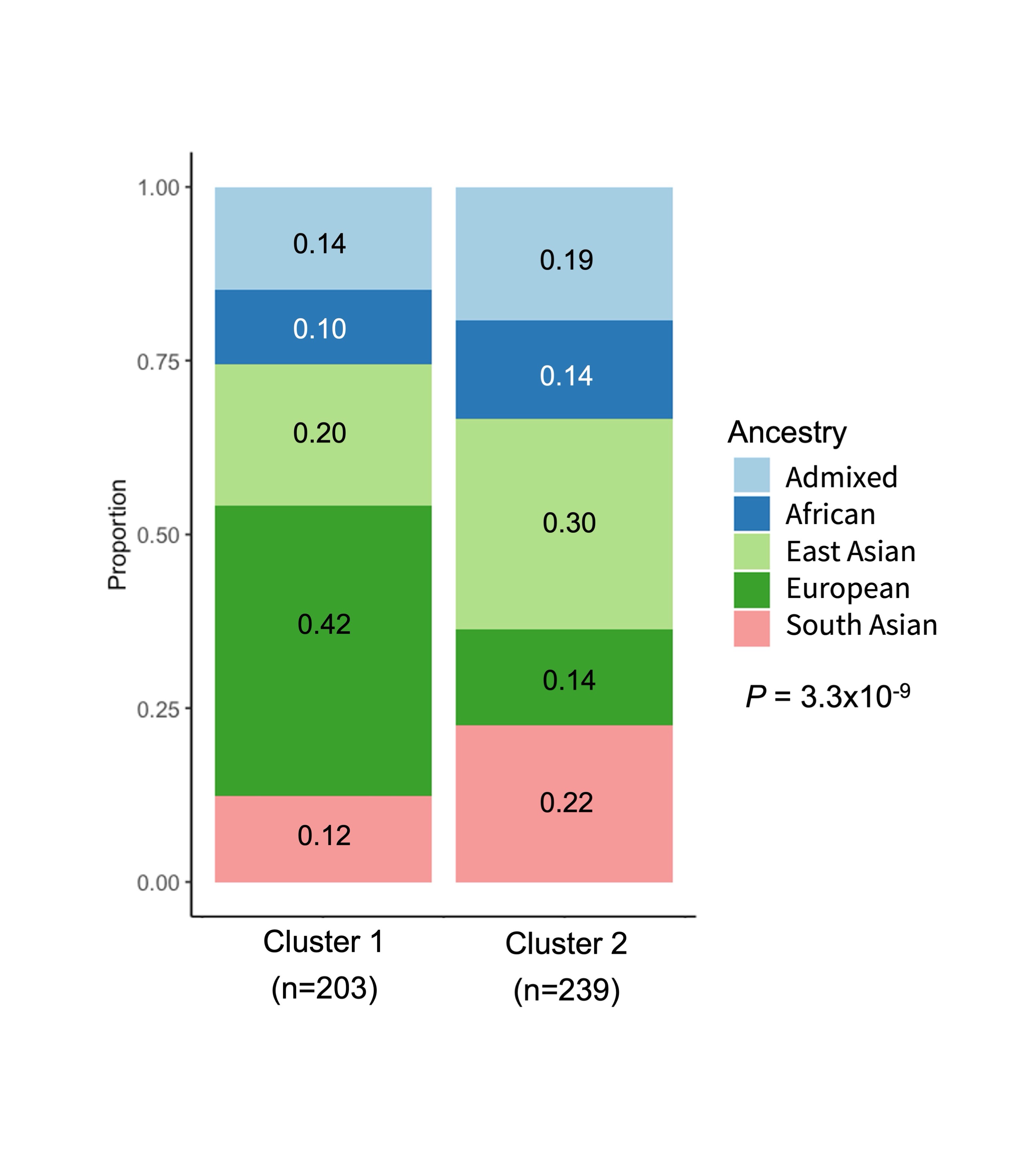
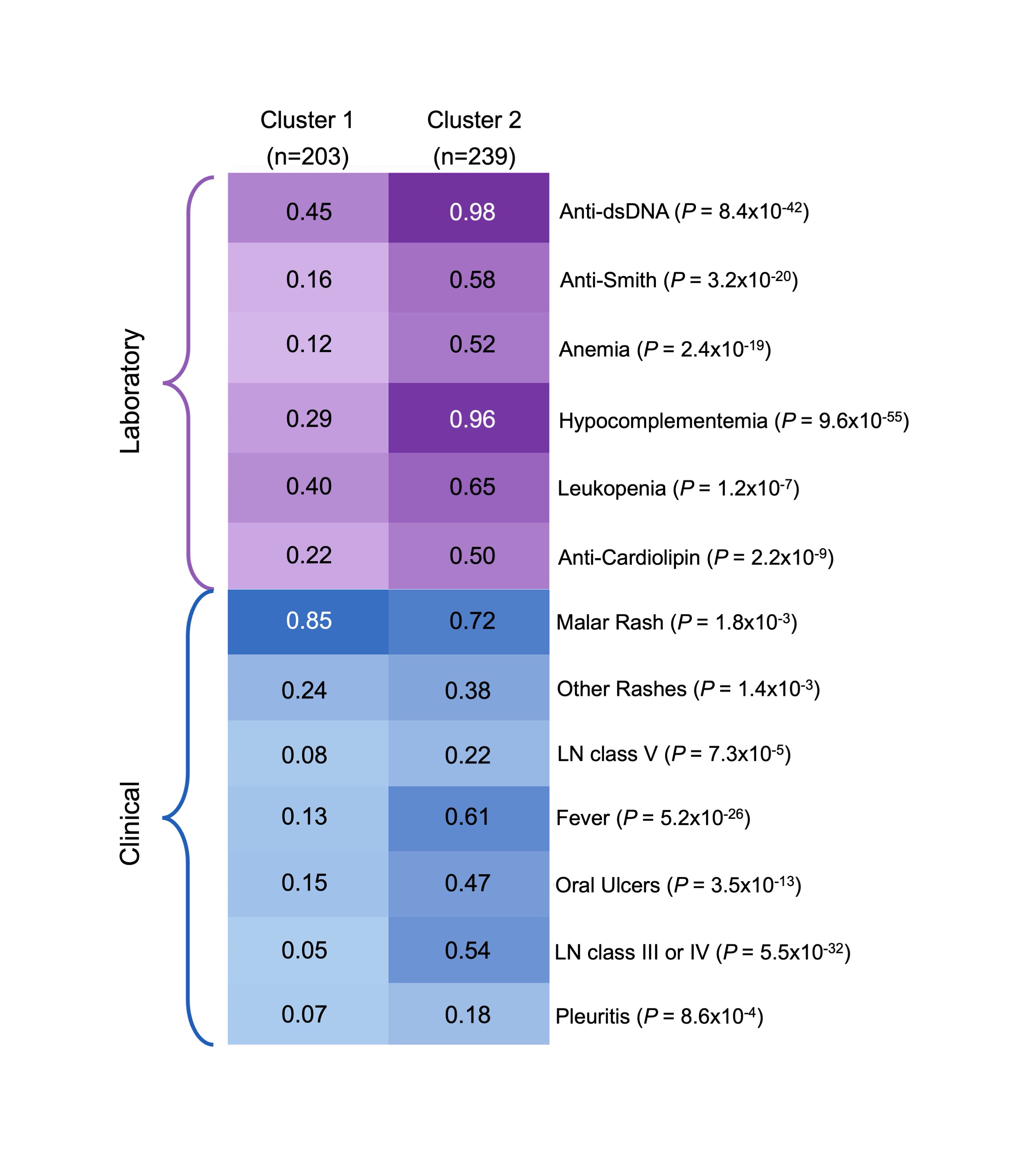
Results: Our cohort included 442 cSLE patients. 83% were female and the median age of SLE diagnosis was 13.6 years (Q1-Q3:12.0-15.8). Our cohort was primarily composed of patients of European (27%) and East Asian (26%) ancestry. SNF identified 2 clusters. The majority of patients in cluster 1 (n=203) were of European ancestry (42%) and cluster 2 (n=239) had a high prevalence of patients of East Asian (30%) and South Asian (22%) ancestry (P=3×10-9; Figure 1). Patients in cluster 2 had a higher prevalence, and earlier onset of class III/IV lupus nephritis, hypocomplementemia, fever, anti-dsDNA and anti-Smith antibodies compared to patients in cluster 1 (P< 1×10-7; Figure 2). The risk of developing 12 cSLE manifestations at any point in time was higher in patients in cluster 2 compared to cluster 1 (HR >1.4;P<4×10-3). SLE PRSs were not associated with cluster membership (Non-HLA SLE PRS: OR 0.9, 95% CI 0.8-1.2; P=0.5; HLA SLE PRS: OR 1.3, 95% CI 0.8-2.0;P=0.3).
Conclusion: In a large multiethnic cohort of cSLE patients, we used data-driven methods to identify two cSLE patient clusters. The cluster with more severe disease and earlier onset had a greater proportion East Asian and South Asian patients compared to the cluster with milder disease. Next steps include an HLA-wide analysis of cluster membership.
To cite this abstract in AMA style:
Chan N, Gold N, Levy D, Knight A, Silverman E, Dominguez D, Ng L, Erdman L, Hiraki L. Identifying Homogeneous Endophenotypes in Childhood-Onset Systemic Lupus Erythematosus with Similarity Network Fusion [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/identifying-homogeneous-endophenotypes-in-childhood-onset-systemic-lupus-erythematosus-with-similarity-network-fusion/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identifying-homogeneous-endophenotypes-in-childhood-onset-systemic-lupus-erythematosus-with-similarity-network-fusion/