Session Information
Session Type: Abstract Session
Session Time: 2:00PM-3:30PM
Background/Purpose: Genome-Wide Association Studies (GWAS) have unveiled over 1000 risk variants for lupus, predominantly situated in non-coding genomic regions. Their functional roles, especially their potential impacts on the dysregulated type I interferon pathway inherent in lupus, remain largely uncharted. To bridge this knowledge gap, our study employs CRISPR/Cas9 technology, enabling a deeper exploration into the functional implications of these risk loci.
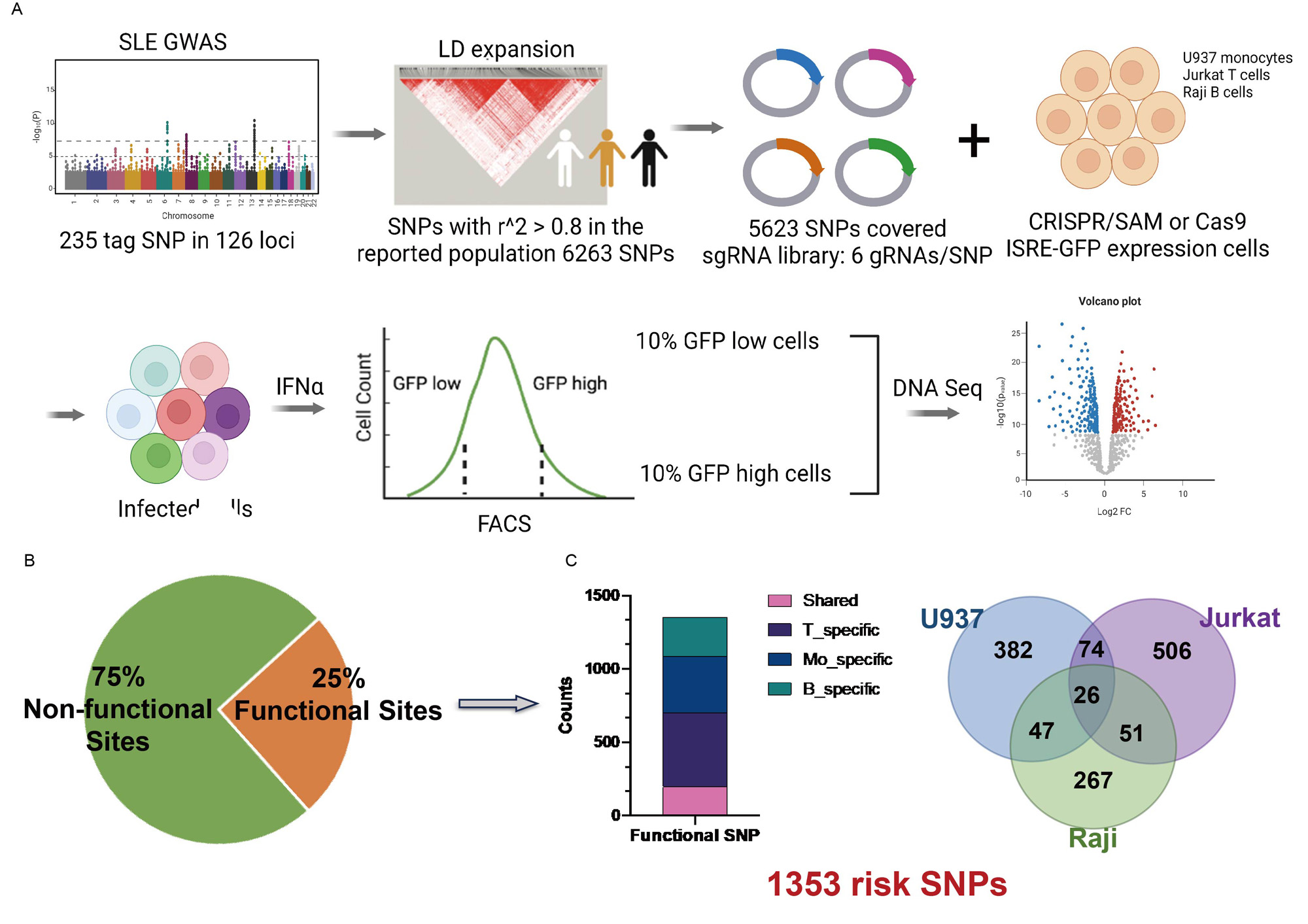
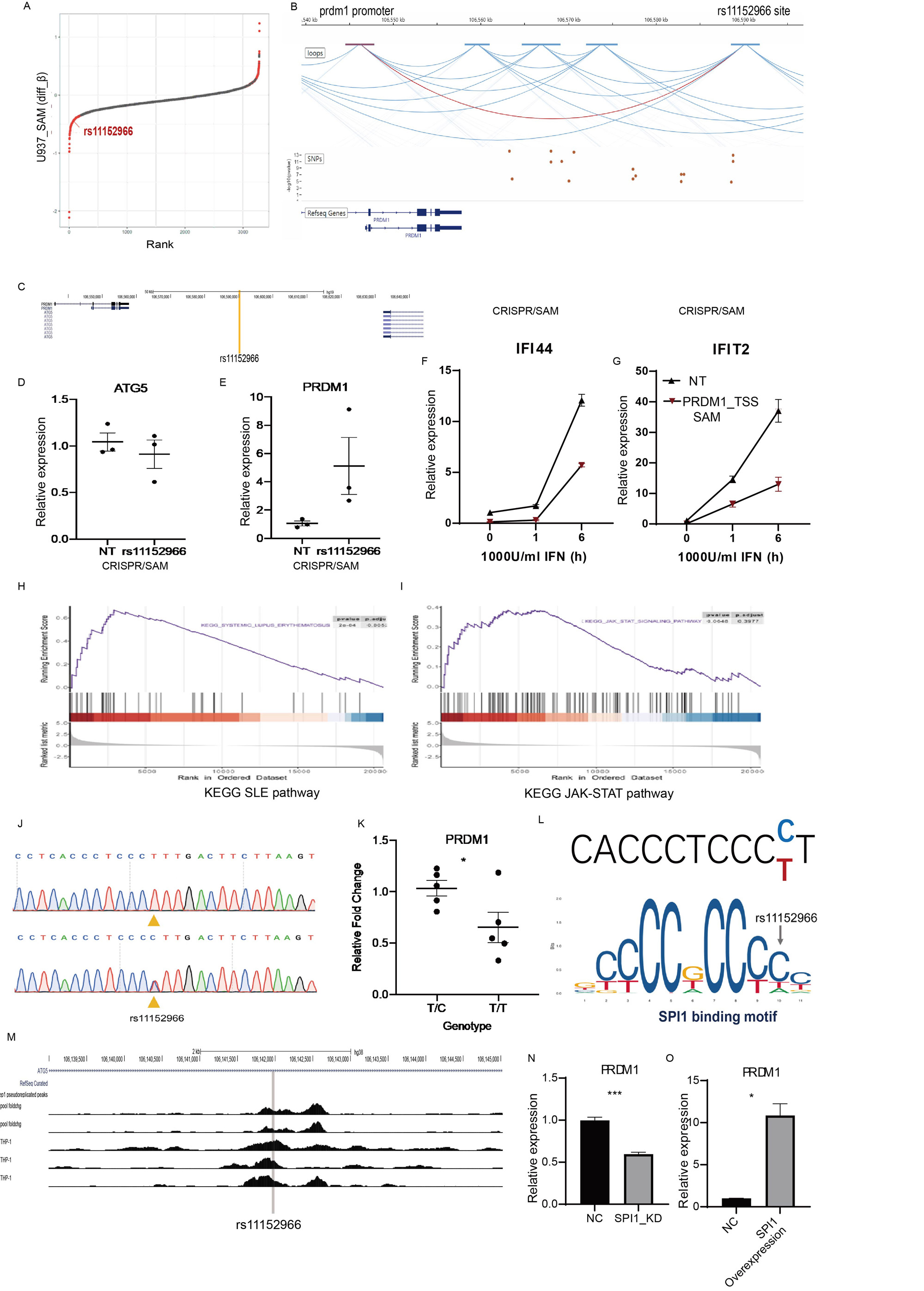
Methods: Our novel strategy entailed CRISPR-based screening for functional variants that regulate the type I interferon pathway. We collected all known SLE-associated SNPs and expanded them through linkage disequilibrium in the population where the SNP was reported. We designed an sgRNA library covering these SLE-associated SNPs and developed a cellular model using U-937, Jurkat, and Raji cells that stably express the CRISPR/Cas9-ISRE-GFP and CRISPR/SAM-ISRE-GFP systems. Post-transduction with the sgRNA lentiviral library, we sorted cells based on GFP intensity and extracted and sequenced DNA from these sorted cells. The functional variants were identified and characterized using multi-omics analysis (eQTL data, HiC data, RNA-sequencing data). We validated the function of the rs11152966-located region and identified PRDM1 as the target gene of rs11152966 through integration of advanced technologies like capture-HiC, CRISPR-SAM, and prime editing technology.
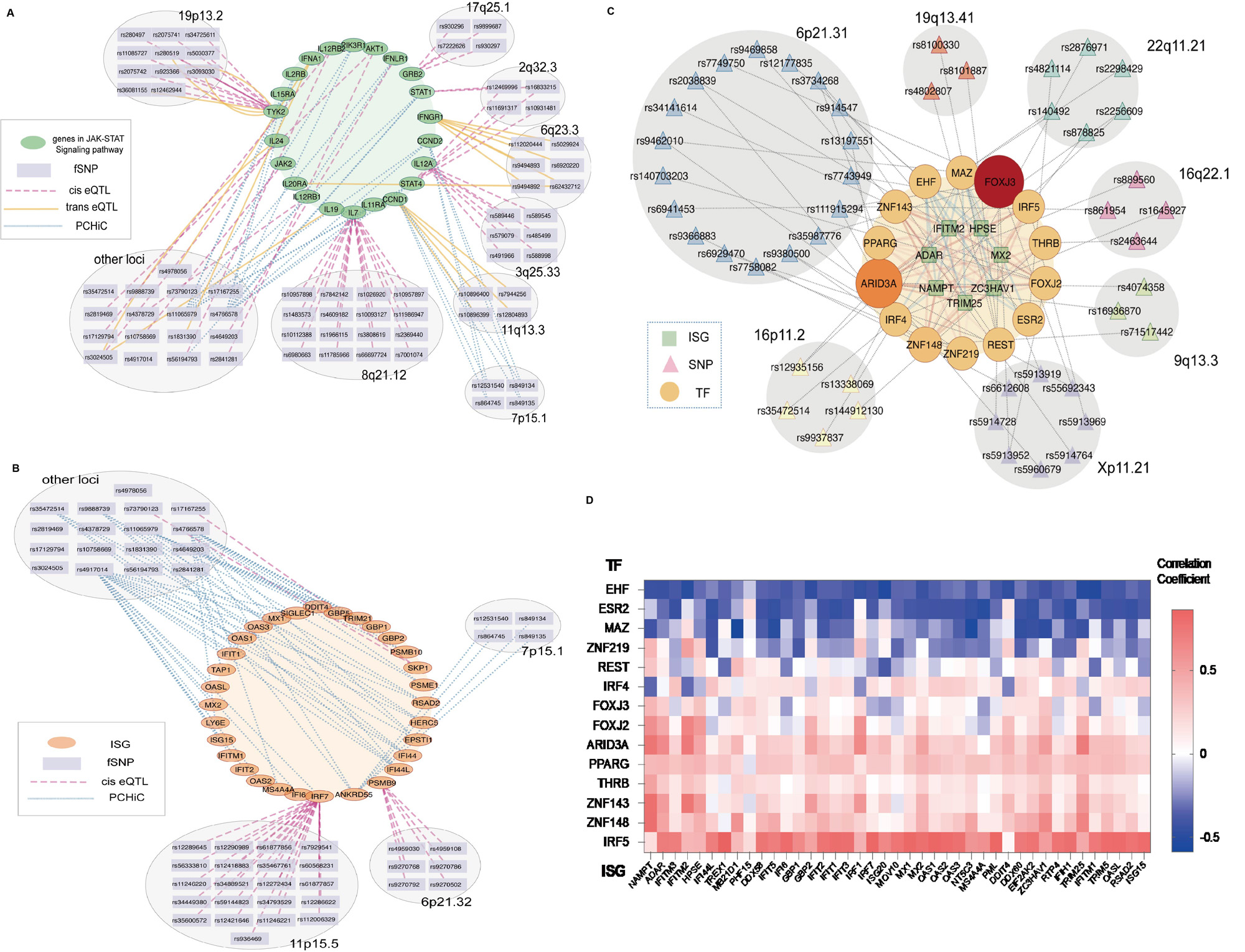
Results: We’ve characterized the comprehensive landscape of functional SNPs related to lupus that regulate the Type I interferon pathway. Notably, we discovered that the majority of these functional variant regions are cell type-specific .Utilizing the CRISPR/Cas9 and CRISPR/SAM systems effectively, we found these tools to have complementary benefits in the identification of these functional variants. Furthermore, these functional variant regions were found to be associated with the expression of critical regulators within the JAK-STAT pathway and interferon-stimulated genes (ISGs). Importantly, we observed a correlation between the expression of transcription factors disrupted by these functional variants and the expression of ISGs in SLE patients. One standout finding was the negative regulatory effect on the Type I interferon pathway upon activation of the region containing the SNP rs11152966. This region, inclusive of the SNP, forms a gene loop with the PRDM1 promoter, thereby managing PRDM1 expression. Additionally, we provided evidence that PRDM1 acts as a novel negative regulator of the Type I interferon pathway. Lastly, our research revealed a link between the presence of the risk allele T and a reduction in PRDM1 expression. This reduced expression appears to be due to the allele T’s effect on the binding of the transcription factor SPI1.
Conclusion: Our study leveraged advanced gene-editing tools to elucidate the landscape of functional SNPs in lupus, revealing their cell type-specific regulation of the Type I interferon pathway and their influence on key genes in the JAK-STAT pathway. Notably, we highlighted the significant role of SNP rs11152966 and its target gene PRDM1 in regulating the interferon pathway in lupus.
To cite this abstract in AMA style:
Hou G, Zhu X, Zhang Y, Cheng Z, Shen N. Global Identification of Lupus Genetic Risk Variants Facilitating the Type I Interferon Pathway Through CRISPR-based Genomic Screening [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/global-identification-of-lupus-genetic-risk-variants-facilitating-the-type-i-interferon-pathway-through-crispr-based-genomic-screening/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/global-identification-of-lupus-genetic-risk-variants-facilitating-the-type-i-interferon-pathway-through-crispr-based-genomic-screening/