Session Information
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: Despite recent advances, the current state of SLE therapeutics remains largely empirical with existing immunosuppressive treatments failing to induce remission in over 40% of patients. There is currently a paucity of tools that would predict response to treatment and guide therapeutic choices.
Methods: Whole blood transcriptome samples were obtained from 95 patients with moderate to severe SLE at baseline, 1 month and 6 months after initiation of treatment with cytotoxic agents (cyclophosphamide, mycophenolate mofetil), mycophenolate mofetil/anti-CD40 antibody, rituximab or belimumab. Disease activity was assessed using the SLEDAI-2K. Response to treatment was defined as achievement of Low Disease Activity State (LLDAS) or remission at 6 months. Differentially expressed genes (DEGs) were identified using the DEseq2. Weighted correlation network analysis (WGCNA) was applied to detect modules of co-expressed transcripts. Abundances of cell types were assessed by CIBERSORTx.
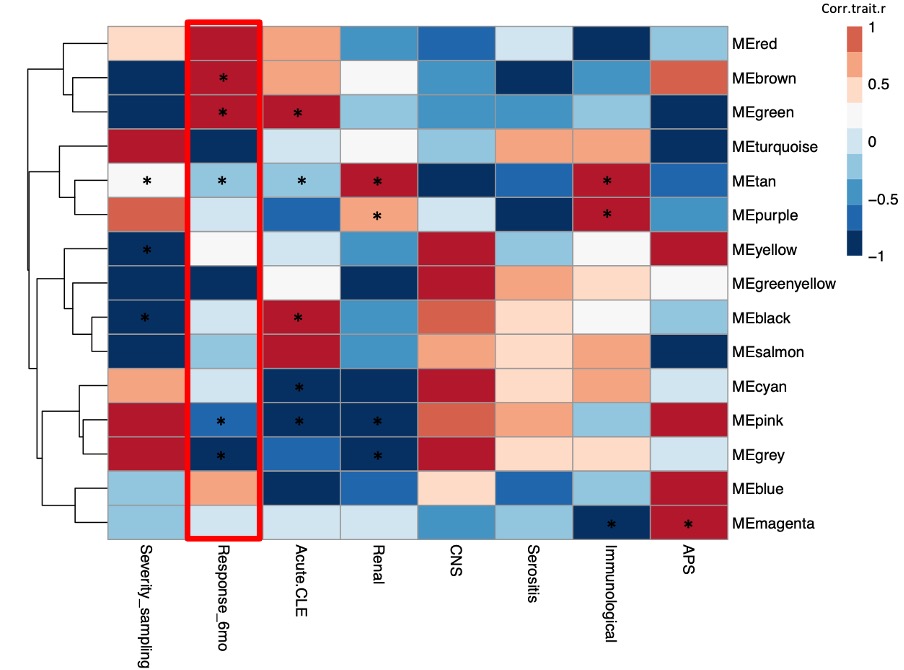
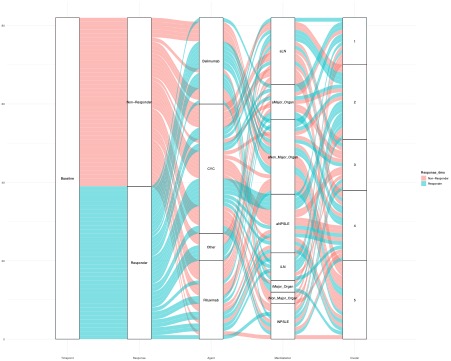
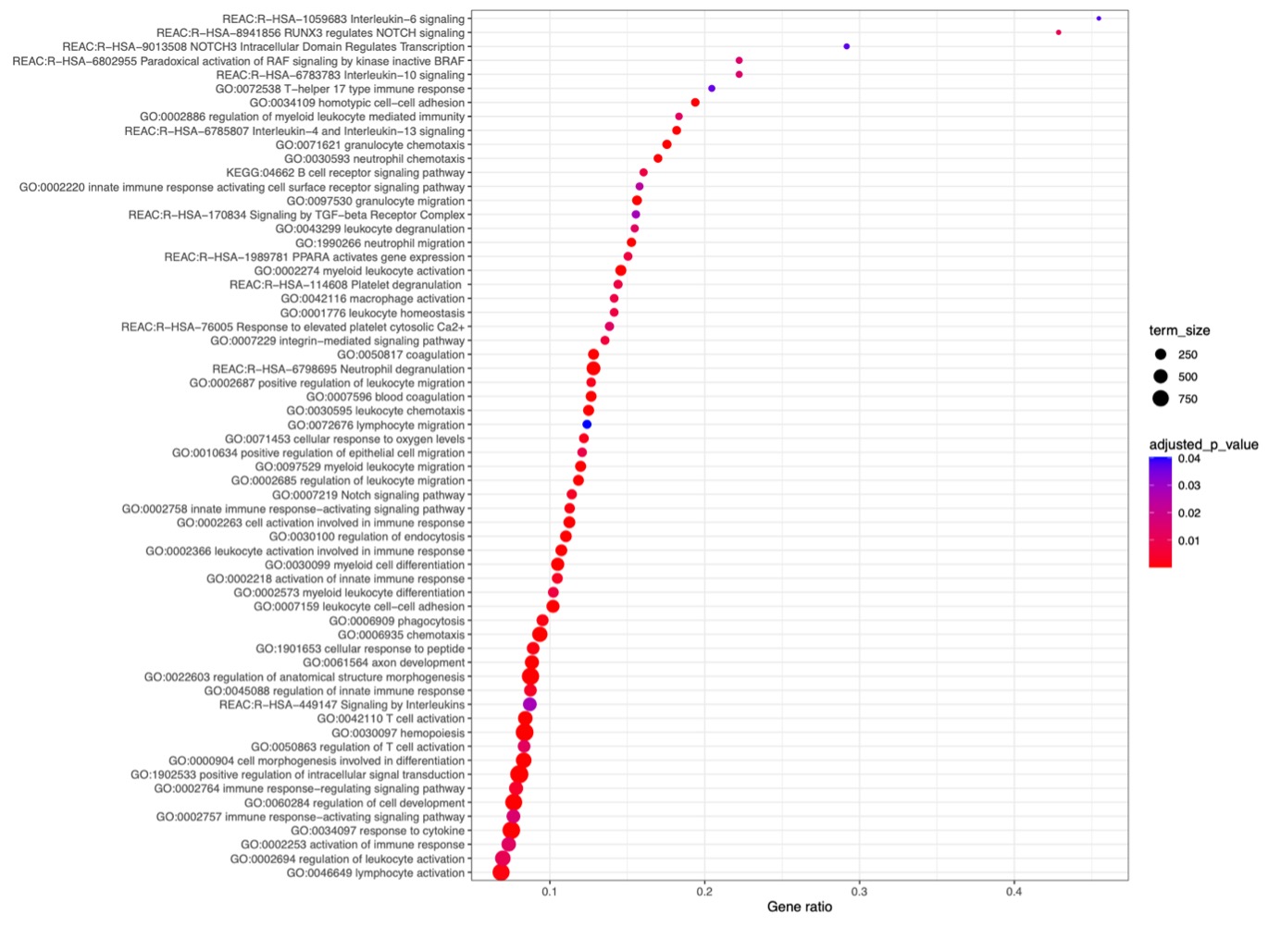
Results: Out of 95 patients the majority were women (93.7%) with a mean [SD] age at SLE diagnosis of 42.9 [13.6] years and a mean disease duration [SD] at sampling of 5.1 [7.2] years. At baseline, mean SLEDAI [SD] was 9.4 [5.7] and active lupus nephritis (LN) was the most common treatment indication (24%), followed by active neuropsychiatric SLE (NPSLE) (20%). Cyclophosphamide was the most frequently used immunosuppressive agent (n=46), followed by belimumab (n=24) and rituximab (n=21). Forty-three patients responded to treatment. WGCNA-generated modules (Figure 1.) that positively correlated with response to treatment at 6 months (p = 0.005, p = 0.04) were enriched in biological processes that included type I interferon, pattern recognition receptor signaling, and neutrophil degranulation. Unsupervised clustering of the baseline samples, based on the module eigengene, revealed a respondent rich patient subset (Cluster 2), which was characterized by high prevalence of active LN (Figure 2.). The transcriptional landscape of “resistant” disease was dominated by disturbances related to p53-signaling pathway. Upregulation of pathways linked to neutrophil degranulation, Th17 responses, IL6 mediated responses, TGF-beta, PPARA, and NOTCH signaling differentiated the one-month-transcriptional profile of responders compared to non-responders (Figure 3.).
Conclusion: Baseline molecular fingerprints related to innate immunity correlated with 6-month response to treatment in SLE. Aberrances linked to p53-signaling decisively shaped the transcriptional signature of “resistant” disease.
To cite this abstract in AMA style:
garantziotis P, Moysidou G, doumas s, nikolopoulos D, Flouda S, kapsala N, Filia A, Sentis G, Fanouriakis A, Bertsias G, Dimitrios B. Baseline Innate Immunity Transcriptional Signatures Act as Predictors of Response to Immunosuppressive and Biologic Treatments in Systemic Lupus Erythematosus While Disturbances Linked to p53-signaling Define “Resistant” Disease [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/baseline-innate-immunity-transcriptional-signatures-act-as-predictors-of-response-to-immunosuppressive-and-biologic-treatments-in-systemic-lupus-erythematosus-while-disturbances-linked-to-p53-signalin/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/baseline-innate-immunity-transcriptional-signatures-act-as-predictors-of-response-to-immunosuppressive-and-biologic-treatments-in-systemic-lupus-erythematosus-while-disturbances-linked-to-p53-signalin/