Session Information
Date: Monday, November 8, 2021
Title: Systemic Sclerosis & Related Disorders – Clinical Poster II (1364–1390)
Session Type: Poster Session C
Session Time: 8:30AM-10:30AM
Background/Purpose: Systemic sclerosis (SSc) patients can be divided into four molecular subsets (inflammatory, fibroproliferative, limited and normal-like) identifiable with gene expression in skin and peripheral blood cells (PBCs). The inflammatory subset typically associated with the presence of inflammatory lymphocyte and innate immune cell infiltrates. SSc participants assigned to the inflammatory subset showed significant improvement in the Abatacept Systemic SclErosis Trial (ASSET). In order to provide a fast and more accessible inflammatory subset predictor, we trained a deep neural network to identify these participants using the images of immunohistochemically (IHC)-stained skin biopsies.
Methods: 164 whole-slide microscopy images were obtained from the ASSET clinical trial. Molecular-subset labels for each whole-slide image were obtained using a previously reported support vector machine (SVM) classifier applied to paired gene expression profiles from the same skin biopsy. A blinded rheumatologist (RL) scored the extent of inflammation and fibrosis in each biopsy. Images were split into training, validation, and test set (70%/15%/15%) with a focus on predicting the inflammatory subset. Small patches were generated using a sliding-window approach and used to train a convolutional neural network (CNN) to infer the subset of the given biopsy. Spearman correlations between predicted probabilities and clinical inflammation scores were examined to validate our model.
Results: A CNN was trained and validated on small patches generated from 140 whole-slide images. The model was evaluated using a hold-out test set of 24 whole-slide images. The final model identifies participants with inflammatory gene signature with 83.33% accuracy (F1-score of 0.75) when data sets are split-by-patient, and 91.30% accuracy (F1-score of 0.9) when data sets are split-by-biopsy. Visualization of patches and predicted decisive regions provided insights on important cell types contributing to the molecular subsets. Areas of the image important for inflammatory subset prediction were identified and visualized. The predicted inflammatory probabilities were significantly correlated with clinical inflammation scores, as assessed by RL.
Conclusion: We trained a CNN that classifies SSc patients into the inflammatory molecular subset using standard IHC images of skin biopsies. This provides a method that is less costly than genomic assays to identify these individuals and can be applied to evaluate retrospective data where genomic data were not collected but for paraffin embedded blocks or IHC-stained images are available.
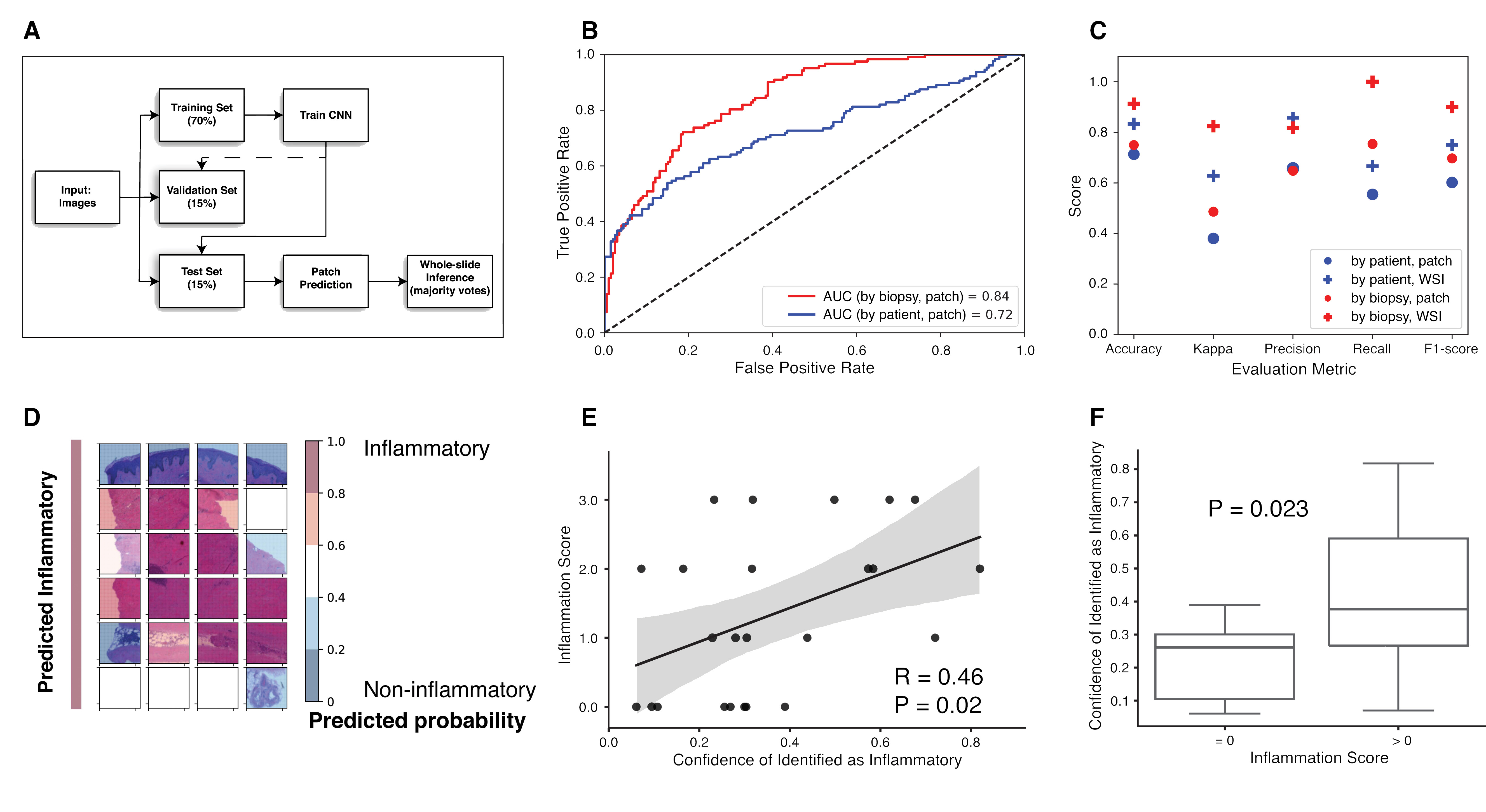 Figure 1. (A) Schematic depiction of the dataset partitioning. (B) Receiver operating characteristic (ROC) curves and their area under the curves (AUCs) on the test set. (C) Models’ performance on the test set for models trained on splitting by patient or by biopsy datasets, patch-based and post aggregation whole-slide level. (D) Visualization of patches in a representative true inflammatory slide by overlapping a colored heatmap based on predicted probability. (E) Correlation of confidence scores for prediction as inflammatory subset and clinical inflammation scores. (F) Whole-slide images with inflammation showed significantly higher confidence scores for prediction as inflammatory subset.
Figure 1. (A) Schematic depiction of the dataset partitioning. (B) Receiver operating characteristic (ROC) curves and their area under the curves (AUCs) on the test set. (C) Models’ performance on the test set for models trained on splitting by patient or by biopsy datasets, patch-based and post aggregation whole-slide level. (D) Visualization of patches in a representative true inflammatory slide by overlapping a colored heatmap based on predicted probability. (E) Correlation of confidence scores for prediction as inflammatory subset and clinical inflammation scores. (F) Whole-slide images with inflammation showed significantly higher confidence scores for prediction as inflammatory subset.
To cite this abstract in AMA style:
Yuan Y, Lafyatis R, Gudjonsson J, Khanna D, Whitfield M. A Deep Neural Network Classifier to Identify Inflammatory Systemic Sclerosis Patients from Histological Images [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/a-deep-neural-network-classifier-to-identify-inflammatory-systemic-sclerosis-patients-from-histological-images/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-deep-neural-network-classifier-to-identify-inflammatory-systemic-sclerosis-patients-from-histological-images/
