Session Information
Date: Saturday, November 7, 2020
Title: SLE – Treatment Poster I
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Atacicept, a dual inhibitor of the B lymphocyte stimulator and a proliferation-inducing ligand (APRIL), has been associated with a reduction of flares in the Phase II/III APRIL-SLE study (NCT00624338) and disease improvement in a subset of patients (pts) with baseline high disease activity (HDA; SLEDAI-2K ≥10) in the Phase IIb ADDRESS II study (NCT01972568). Using immune cell deconvolution of gene expression data, we previously distinguished two pt clusters in the APRIL-SLE study characterized by different rates of flare in the placebo (PBO) group [1]. We proposed an analysis of ADDRESS II to confirm the differential treatment effect observed using baseline gene expression in the APRIL-SLE study.
Methods: In a prior study of APRIL-SLE samples, a cell deconvolution algorithm [2] was used to assess 17 immune cell subsets in gene expression data from mRNA extracted from whole blood at baseline (N=105). Unsupervised clustering revealed five clusters of pts (P1–P5). In two clusters (P1,3), PBO-treated pts, when compared with atacicept-treated pts, had few new BILAG A or B flares. In the other three clusters (P2,4,5), PBO-treated pts had high flare rates. The P2,4,5 subset includes pts with high plasmablasts, high B cells and low neutrophils, or high activated natural killer cells and high activated dendritic cells. Using the cellular modules that defined these clusters, we developed an algorithm to distinguish P1,3 from P2,4,5 and applied it to the ADDRESS II mITT and HDA populations with gene expression data at baseline. Treatment arms in the two resulting subsets were compared using SLE Responder Index (SRI)-4, SRI-6, BILAG-based Combined Lupus Assessment (BICLA), and low disease activity (SLEDAI-2K ≤2).
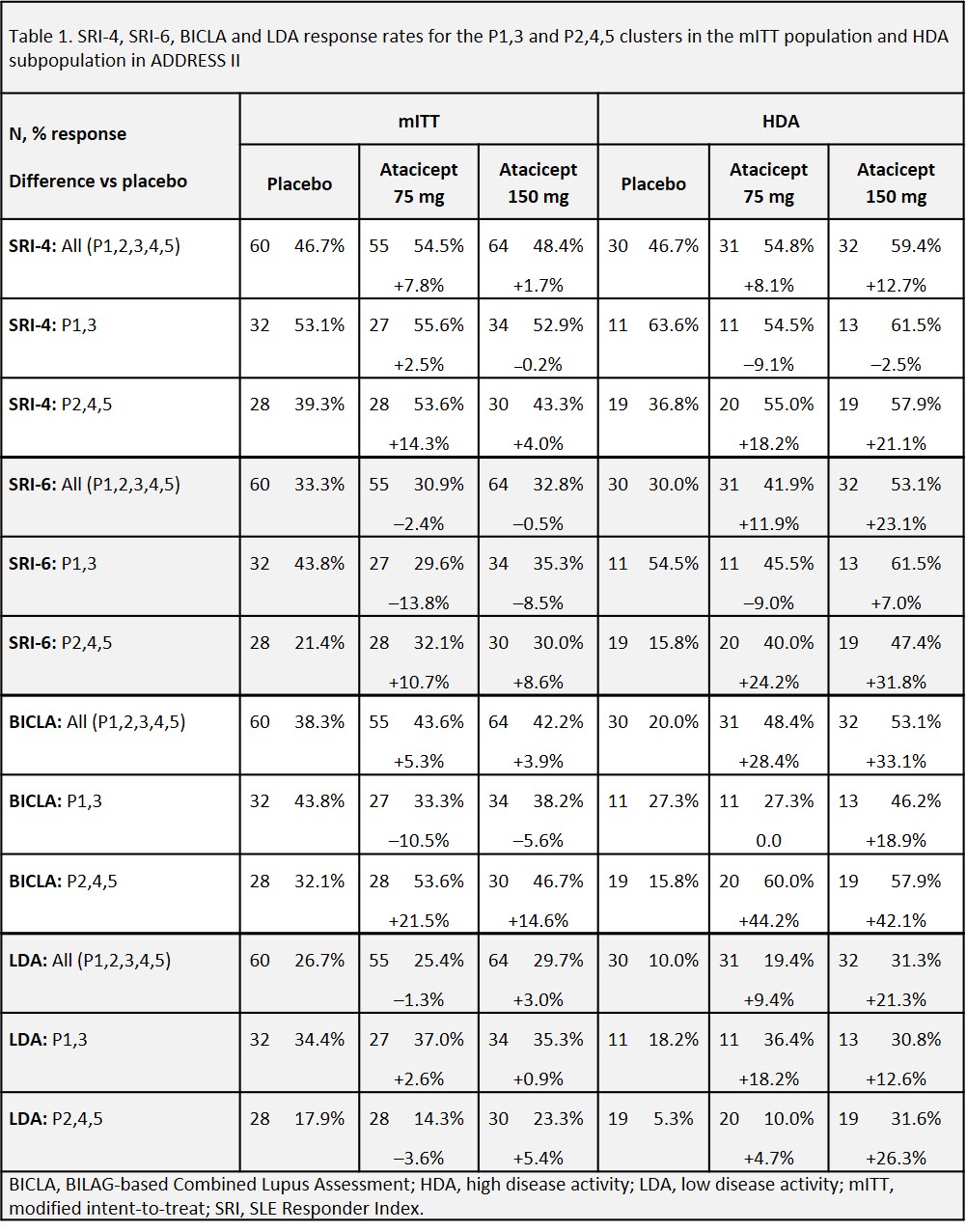
Results: In ADDRESS II, 179 pts (58.5%) of the mITT and 93 pts (59%) of the HDA subpopulation had RNAseq expression data; their clinical response rates were similar to the overall mITT and HDA populations. The algorithm assigned 93 mITT pts (52%) to the P1,3 subset and 86 (48%) to the P2,4,5 subset. For HDA pts, 35 (38%) were assigned to P1,3 and 58 (62%) to P2,4,5. The P2,4,5 subset was confirmed to have lower PBO response rates and higher treatment difference for atacicept compared to the P1,3 subset as measured by SRI-4, SRI-6, and BICLA in both mITT and HDA populations. (Table 1).
Conclusion: This exploratory analysis of the ADDRESS II trial demonstrated greater treatment effect of atacicept in a subset of pts identified by an algorithm derived from unsupervised clustering of whole blood gene expression in a different atacicept study. The observation that better response differences were found in pts with high B cells or high plasmablasts is consistent with atacicept’s proposed mechanism of action.
- Samy et al. 2019 Lupus Science & Medicine;6(Suppl 1):A158-A; 2. Abbas et al. 2009 PLOS ONE;4(7):e6098
To cite this abstract in AMA style:
Merrill J, Studham M, Morand E, Aydemir A, Vazquez Mateo C, Rolfe A, Kao A, Townsend R. Identifying an SLE Patient Cluster with Greater Treatment Effect: Immune Cell Deconvolution of Gene Expression in Two Atacicept Phase II Studies [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/identifying-an-sle-patient-cluster-with-greater-treatment-effect-immune-cell-deconvolution-of-gene-expression-in-two-atacicept-phase-ii-studies/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identifying-an-sle-patient-cluster-with-greater-treatment-effect-immune-cell-deconvolution-of-gene-expression-in-two-atacicept-phase-ii-studies/