Session Information
Session Type: Poster Session (Tuesday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Gout encompasses acute arthritis flares mediated by innate autoinflammatory responses to urate crystals, chronic granulomatous tophi, and synovitis promoting bone erosion and soft tissue damage. Here, we probed gout mediators in the circulating mononuclear leukocyte (PBMC) DNA methylome.
Methods: PBMC DNA converted by Na+ bisulfite modification was hybridized to Illumina Infinium MethylationEPIC BeadChip arrays. Methylation data were processed with ChAMP in R. Our North American (NA) gout cohort (n=16), sampled between flares, all met ACR/EULAR criteria, and well-controlled urate overal. Healthy controls (n=14) were matched for age, gender, and BMI. Obese New Zealand (NZ) Maori, enriched in hypertension and diabetes, provided an independent validation cohort (n=13 gout; n=16 no gout). Transcription Factor (TF)-gene networks were built and combined from B and T cells, CD14+ monocytes, CD4+ and CD8+ T cells.
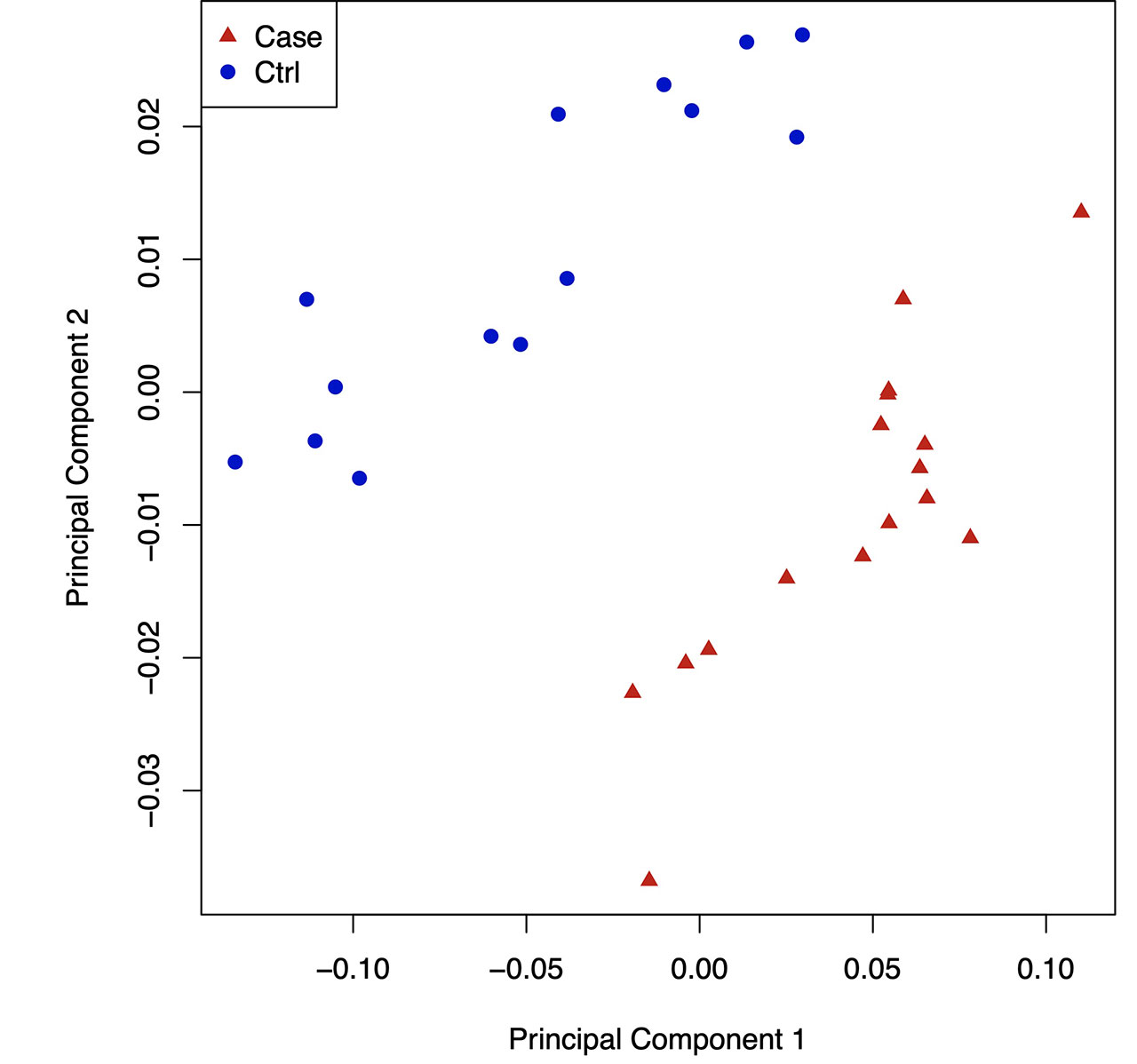
Results: By hierarchical clustering and PCA, gout DNA methylome signatures were significantly separable from controls in both the NA cohort (Fig. 1,2), and NZ validation cohort. Most ( >85%) differentially methylated loci (DML) in gout were hypo-methylated in open sea regions, and gene body hypomethylation (which inhibits gene expression) was prominent. Gout DML included 23 from ~125 genetic risk loci from past GWAS (eg, urate transporter SLC2A9 , B2 kinin receptor, and IL-23R , which bridges innate and adaptive immunity, and promotes granuloma formation). Numerous KEGG signaling pathways, but not the NLRP3 pathway, were enriched in DML (eg, circadian entrainment, and TRP ion channel signaling (which transduces pain in nerve fibers, and also activates phagocytes)), aligned with severe pain and nocturnal onset of background gout flares. We saw differential DNA methylation of myeloid cell signaling and function pathways (eg, FcgammaR phagocytosis, chemokine signaling, adhesion, PI3K-AKT, HIF-1), nutritional biosensing by NF-kB and NLRP3 inhibitor AMPK, and choline metabolism, which promotes gouty inflammation by modulating AMPK and mitophagy. In gout, STAT2 , IRF1 , nuclear factor of activated T cells (NFATC2), and major adaptive immunity pathways (T and B cell receptor signaling, Th17 differentiation) were differentially methylated. By far, most overlaps between gout cohorts were in transcription factors (TFs) with motifs enriched in hypo-methylated sequences, and enriched KEGG pathways based on TFs. Strikingly, DMLs of NFATC2 , which blunts NF-kB hyperactivation, and myocyte enhancer factor 2C MEF2C , which controls myeloid switching to granulocytopoiesis, were among 10 TFs propagated in all PBMC TF networks in both gout cohorts (Fig. 3).
Conclusion: A PBMC DNA methylome signature reflected trained immunity in gout, especially imprinting signaling and transcriptional pathways, and involving adaptive immunity. Such changes, and differential TF-gene network DNA methylation in all PBMCs, were in contrast to lack of NLRP3 pathway imprinting. The PBMC DNA methylome signature likely helps determine phenotypic outcomes of not only acute gouty arthritis, but also granulomatous tophaceous and chronic inflammatory synovitis, and potentially modulates comorbidities in the disease.
To cite this abstract in AMA style:
Wang Z, Zhao Y, Phipps-Green A, Liu-Bryan R, Ceponis A, Boyle D, Wang J, Merriman T, Wang W, Terkeltaub R. Mononuclear Leukocyte DNA Methylome Imprinting of Networked Signaling and Immunity Regulatory Pathways in Gout [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/mononuclear-leukocyte-dna-methylome-imprinting-of-networked-signaling-and-immunity-regulatory-pathways-in-gout/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/mononuclear-leukocyte-dna-methylome-imprinting-of-networked-signaling-and-immunity-regulatory-pathways-in-gout/