Session Information
Session Type: ACR Abstract Session
Session Time: 4:30PM-6:00PM
Background/Purpose:
Dermatomyositis (DM) is a complex immune-mediated disease resulting in muscle and skin inflammation. Prior studies of gene expression in DM have revealed a type I interferon (IFN) signature, but less is known about alternative immune pathways involved. We conducted a comprehensive gene expression meta-analysis in DM muscle and skin to identify disease-relevant genes and pathways and elucidate a shared immune mechanism across tissues.
Methods: Raw data from 6 publicly available datasets from DM muscle (71 cases, 36 controls) and 2 from skin (77 cases, 22 controls) were downloaded. Data were preprocessed, merged and batch-corrected, creating tissue-specific gene expression matrices. Significance Analysis of Microarrays was used to identify differentially expressed genes (DEG; cutoff FDR p-value < 0.05, FC≧1.3). A hypergeometric test was used to assess overlap between DEGs in skin and muscle. Weighted Gene Coexpression Network Analysis was used to construct muscle and skin networks and identify associations not detected on the single gene level. Disease-associated modules were identified by correlating module eigengene with case/control status. Network preservation was assessed by calculating gene overlap between muscle and skin modules using a hypergeometric test. Hub genes (gene significance >0.2 & module membership >0.8) in each network were identified. Cell type enrichment was performed using xCell.
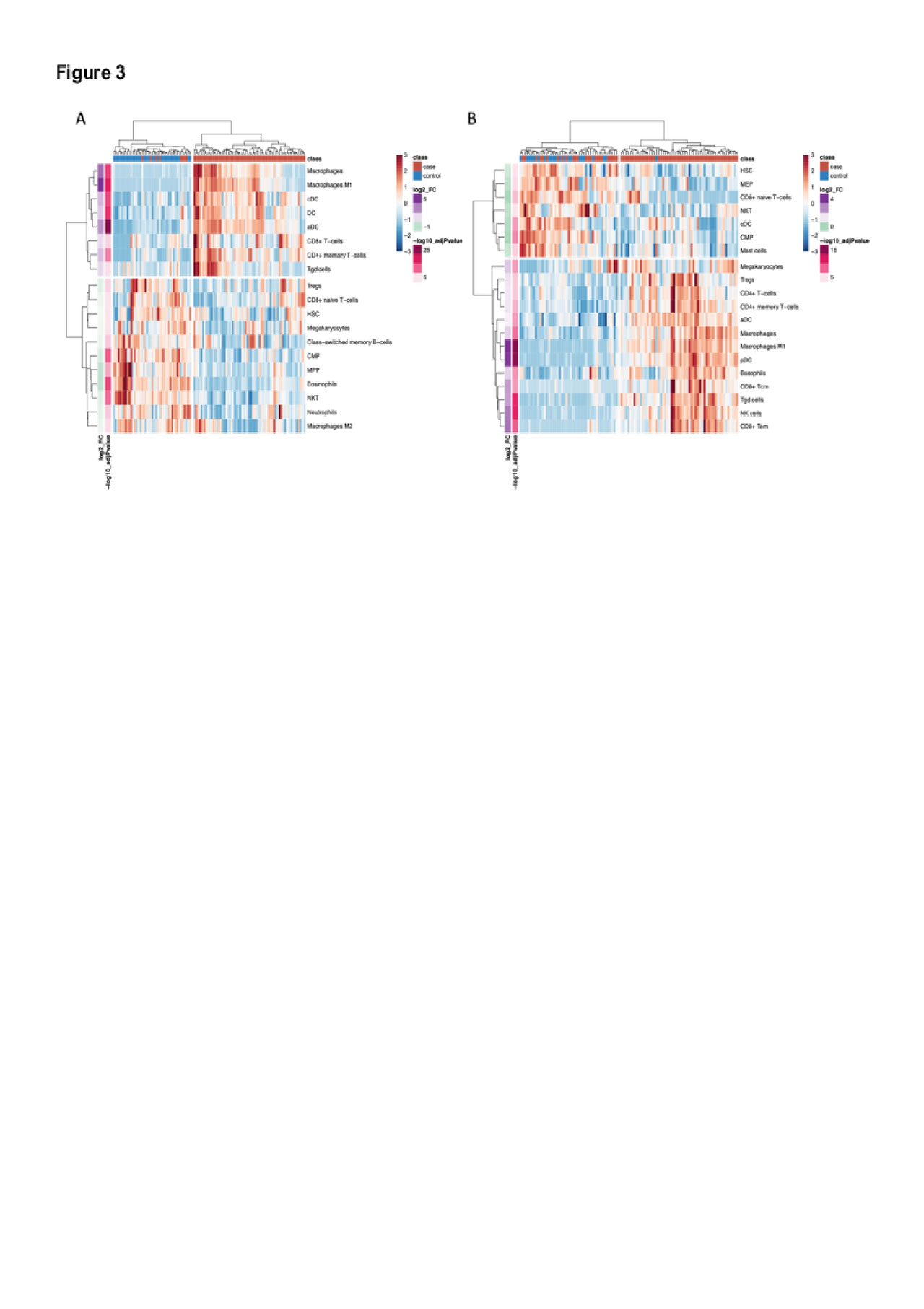
Results: There were 544 significantly DEGs (FC≧1.3, q< 0.05) in muscle and 300 in skin (Fig 1A & 1B). There was significant overlap in upregulated genes across tissues with 94 shared genes (p=6.4xE-78) which were enriched in type I and II IFN signaling and MHC class I antigen processing pathways (Fig 1C & 1D). In a network analysis, we identified 8 significant muscle modules and 7 skin modules either positively or negatively correlated with case/control status (Fig 2A). Modules that were highly correlated with cases also shared significant overlap on the gene level (Fig 2A). These modules were enriched in pathways consistent with the single gene analysis and also in terms related to T cell activation and T cell receptor signaling (Fig 2B). Modules correlated with controls were enriched in processes related to energy metabolism. The top hub genes in each network included genes classically involved in type I IFN (STAT1, MX1, IFI44, ISG15), type II IFN (CXCL10, GBP1), class I MHC antigen processing (HLA-A, -B, -F, B2M, TAP1) and the immunoproteasome (PSMB8, PSMB9). In the cell type enrichment analysis, both tissues were enriched in antigen-presenting cells, including activated dendritic cells and M1 macrophages (Fig 3A & 3B).
Conclusion:
This analysis demonstrates striking similarities in gene expression in target tissues in DM and simultaneous activation of innate and adaptive immune responses. Using an unbiased approach, we show enrichment of type I and type II IFN pathways, MHC class I antigen processing pathways, T cell activation, and antigen-presenting cells. These results suggest type II IFN contributes to the global IFN signature in DM and that altered autoantigen presentation through the class I MHC pathway may be important in disease pathogenesis.
To cite this abstract in AMA style:
Neely J, Rychkov D, Paranjpe M, Waterfield M, Kim S, Sirota M. Gene Expression Meta-Analysis Reveals Commonalities in Gene Activation and Enrichment of Immune Pathways and Cell Types in Dermatomyositis Target Tissues [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/gene-expression-meta-analysis-reveals-commonalities-in-gene-activation-and-enrichment-of-immune-pathways-and-cell-types-in-dermatomyositis-target-tissues/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/gene-expression-meta-analysis-reveals-commonalities-in-gene-activation-and-enrichment-of-immune-pathways-and-cell-types-in-dermatomyositis-target-tissues/