Session Information
Date: Sunday, November 10, 2019
Title: Pediatric Rheumatology – ePoster I: Basic Science, Biomarkers, & Sclerodermic Fever
Session Type: Poster Session (Sunday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Skin inflammation can herald systemic disease manifestations and disease chronicity in juvenile myositis (JM), yet we lack an understanding of the pathogenic mechanisms driving skin inflammation in JM. The objectives of this study were to 1) define transcriptional signatures and biological networks in JM lesional skin, and 2) identify key genes and pathways that differentiate skin disease in JM from childhood-onset SLE (cSLE).
Methods: With IRB approval, we identified JM and cSLE patients who had a skin biopsy performed at either the University of Michigan or Lurie Children’s Hospital of Chicago and verified diagnosis through chart review of clinical findings, labs, imaging and histopathology. We then isolated RNA from formalin-fixed, paraffin-embedded (FFPE) skin biopsy samples of 15 JM (9 lesional, 6 non-lesional), 7 cSLE and 8 control patients (CTL; samples from uninvolved skin removed with nevi excision). Following RNA isolation, we performed microarray analysis using Affymetrix ST 2.1 arrays and significance analysis of microarrays (SAM) to determine differentially expressed genes (DEGs; q-value ≤ 5%) between patient groups, comparing JM lesional (JM_L), JM non-lesional (JM_NL), cSLE and CTL. We utilized Ingenuity Pathway Analysis (IPA) to highlight enriched biological pathways and Genomatix Pathway System (GePS) to characterize regulated genes within biological networks.
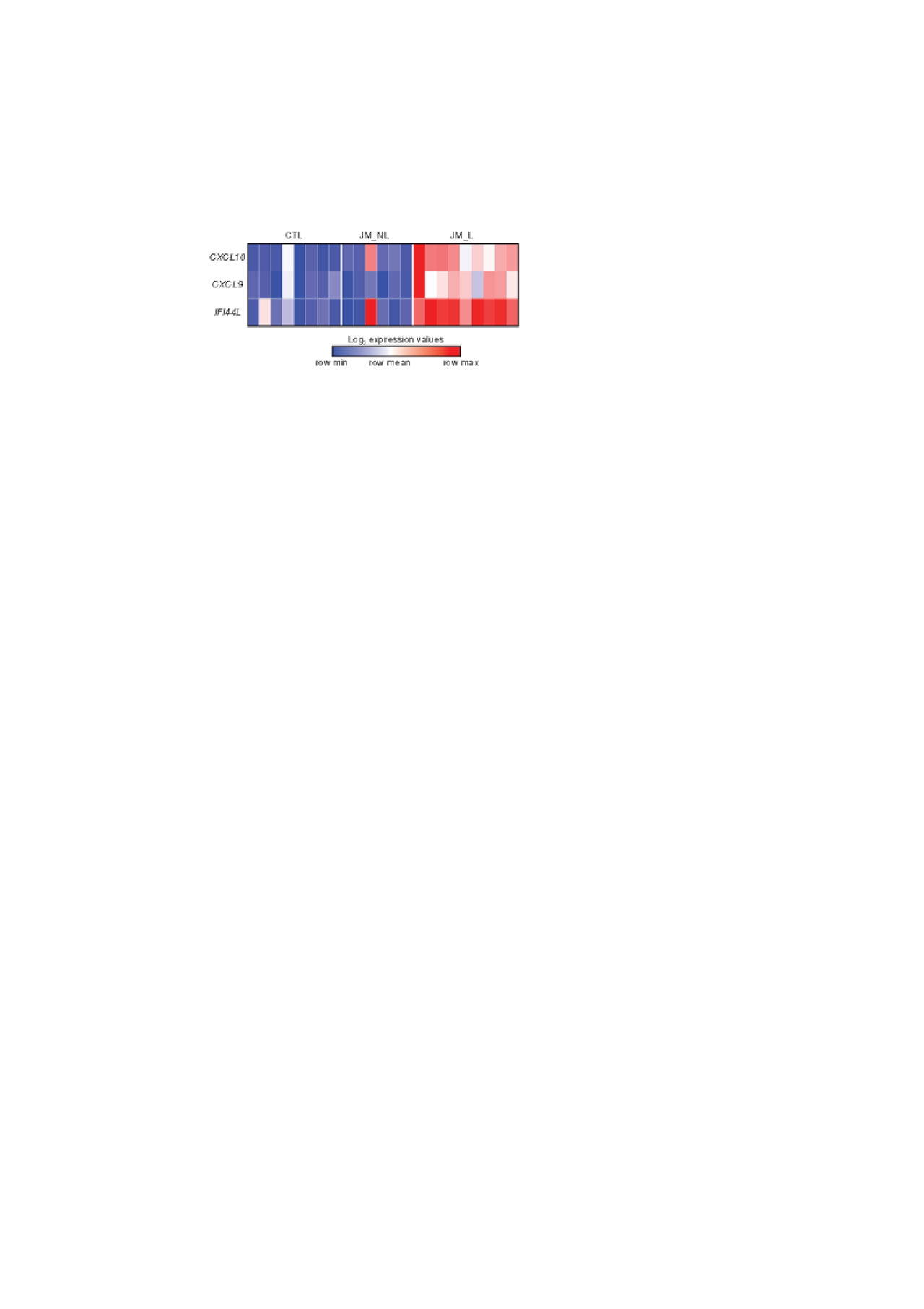
Results: In the JM cohort, 2 patients had amyopathic disease, and 2 lacked characteristic skin findings of Gottron’s papules, with the remaining patients having a classical JDM presentation. Principal component analysis demonstrated that the 2 patients without characteristic skin findings of JDM clustered with other JM_L patients. Comparison of JM_L to CTL skin revealed 221 DEGs, with all but one upregulated in JM_L. The majority of upregulated genes in JM_L were IFN-sensitive, with CXCL10, CXCL9 and IFI44L representing the top three DEGs (FC = 23.2, 13.3, 13.0, q-value = < 0.0001) (Figure 1). IPA revealed IFN signaling as the top canonical pathway and also showed upregulation of antigen presentation, pattern recognition receptors, communication between innate and adaptive immune cells, T cell signaling, the complement system and dendritic cell maturation. The top predicted upstream regulator was IFN alpha (p-value = 7.91×10-88). JM_NL skin had a strikingly different transcriptional signature, both lacking a prominent IFN signal and with downregulation of multiple genes and pathways, including oxidative phosphorylation and protein ubiquitination. JM_L skin shared a highly similar gene expression pattern with cSLE. There were only 28 unique DEGs in JM_L compared to cSLE, whereas cSLE skin had 722 unique DEGs compared to JM_L. Of note, cSLE skin uniquely demonstrated increased expression of IFN gamma relative to CTL.
Conclusion: JM lesional skin demonstrates a striking IFN signature similar to that previously reported in muscle and peripheral blood. Although JM skin has few unique DEGs when compared to cSLE, a lack of upregulated genes specific to cSLE skin may distinguish JM. The origin of the IFN signature in JM skin and its association with clinical phenotype and response to therapy remains to be established.
Heatmap for JDM_ACR abstract_FINAL
To cite this abstract in AMA style:
Turnier J, Berthier C, Tsoi L, Lowe L, Morgan G, Gudjonsson J, Pachman L, Kahlenberg J. Cutaneous Gene Expression Signatures in Juvenile Myositis Reveal a Prominent IFN Signature in Lesional Skin [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/cutaneous-gene-expression-signatures-in-juvenile-myositis-reveal-a-prominent-ifn-signature-in-lesional-skin/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/cutaneous-gene-expression-signatures-in-juvenile-myositis-reveal-a-prominent-ifn-signature-in-lesional-skin/
