Session Information
Session Type: ACR Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Systemic sclerosis (SSc) causes varying degrees of skin fibrosis with varying trajectory. Extent of skin involvement is measured by the modified Rodnan skin score (mRSS), a semi-quantitative clinical method of pinching the skin to determine fibrosis. Previously, a 415-gene signature in skin was identified whose change between baseline and 12 months predicted mRSS at 24 months. We identified a skin gene expression signature at a single time point that predicts mRSS 6 or 12 months later.
Methods: Gene expression profiles were obtained from 260 skin biopsies from 45 SSc patients treated with mycophenolate mofetil (MMF) and 22 healthy controls. Participants were classified into progressing, stable, or regressing groups based on the mRSS change between the baseline skin biopsy and the subsequent timepoint 6 or 12 months later. A multinomial generalized linear model determined the probability of skin trajectory from gene expression profiles and clinical information.
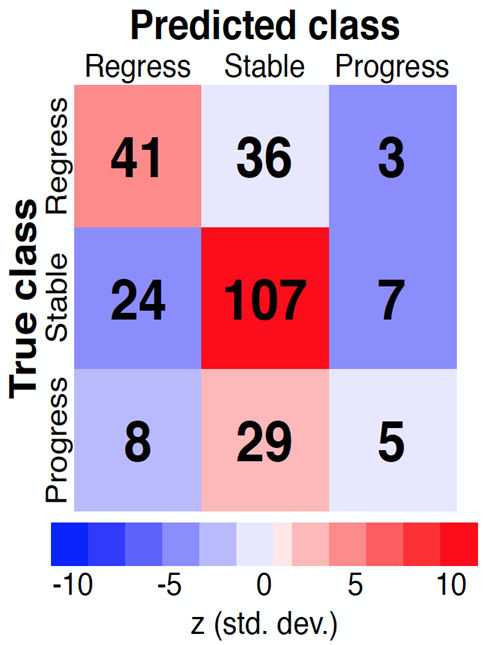
Results: The mRSS regressing group compared to the mRSS progressing and stable groups combined demonstrated 762 differentially expressed genes (823 probes). A multinomial generalized linear model that included genomic and clinical data was developed to predict skin trajectory with 58.8% total accuracy compared to 40% using a random permutation null model. Up-regulated genes in improvers were enriched for extracellular matrix (ECM) metabolism, transforming growth factor-pathway expression, and angiogenesis.
Conclusion: We demonstrate proof-of-principle that significant prognostic information may be obtained from analysis of skin gene expression at a single time point. Further investigation is warranted to determine the generalizability of the model. Future directions include incorporating additional information such as serum protein data and histologic data to increase the predictive accuracy of the model.
Figure 1. Error matrix comparing the performance of the experimental model to a null model. The null model permutes 1000 times yielding a mean prediction accurate to 0.1. The experimental model more accurately predicts patients whose mRSS improves or remains stable compared to the null model.
To cite this abstract in AMA style:
Correia C, Carns MA, Aren K, Hinchcliff M, Mahoney JM. Skin Gene Expression Profiling Predicts Longitudinal Modified Rodnan Skin Score Change [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/skin-gene-expression-profiling-predicts-longitudinal-modified-rodnan-skin-score-change/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/skin-gene-expression-profiling-predicts-longitudinal-modified-rodnan-skin-score-change/