Session Information
Date: Monday, October 22, 2018
Title: Systemic Sclerosis and Related Disorders – Basic Science Poster II
Session Type: ACR Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose:
Localized scleroderma (LS) is a progressive disease of the skin and underlying tissue that causes significant functional disability and disfigurement, especially in developing children. During the active inflammatory stage of the disease, systemic immunosuppressive therapy intervention can halt disease progression and ameliorate disease features such as joint contractures. The existing clinical process for determining when LS is active and susceptible to intervention and when to gradually stop intervention is often convoluted. Commercially available inflammatory serologic markers currently have limited utility in the clinical assessment of active disease. RNA sequencing (RNAseq) technology allows for improved understanding of relevant cellular expression through transcriptome analysis of specific time points during LS disease progression. It also permits the use of RNA extracted from existing paraffin-embedded skin tissue increasing the availability of samples.
Methods:
RNAseq was performed on paraffin-embedded skin (n=15 LS, n=5 pediatric healthy) and RNAlater preserved skin specimens (n=2 LS, n=4 healthy) using the Illumina HTS using TrueSeq Access library preparation and collected through IRB#PRO11060222. Paired end RNA sequencing data was aligned using STAR and analyzed for differential gene expression (DEGs) using DESeq2. Genes were analyzed using DEG cutoffs of log2fold change > ±2.5, adjusted p<0.05, and a false discovery rate (FDR) cutoff of <0.01 for enrichment software (GSEA©). PCA and hierarchical clustering was performed using Cluster 3.0 and Partek® softwares. Clinical subtype data (active/inflammatory vs. stable/disease damage) was applied to these clustering techniques.
Results:
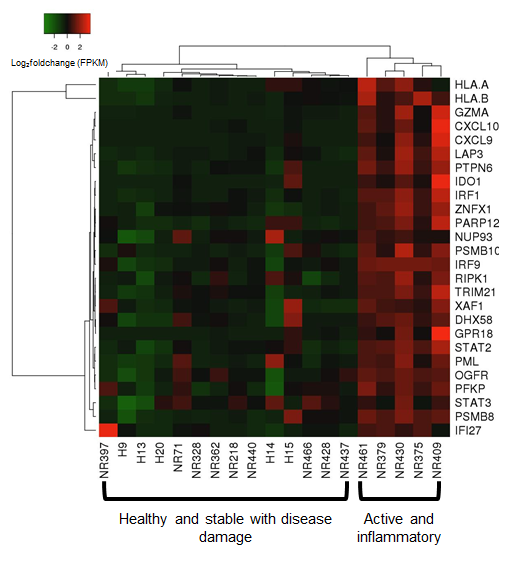
A strong correlation (rS=0.90, p=0.0001) was observed between the comparison of genes expressed between fresh (RNAlater) and paraffinized skin in healthy and LS subjects. When compared to healthy controls, we observed distinct expression of an inflammatory response gene signature (IRGS) composed of IFNγ, IFNα and TNFα associated genes. GSEA© enrichment analysis showed the IRGS, including interferon inducible chemokines such as CXCL9, CXCL10, CXCL11 and IFNγ itself, was more highly expressed in LS patients with more inflammatory lesions (see Figure).
Conclusion:
The use of paraffinized skin for sequencing was proven to be an effective substitute for fresh skin by comparing gene expression profiles. The prevalence of the IFNγ signature in the lesion biopsies of active LS patients indicates these genes are reflecting clinical parameters and provide insight into disease propagation, possibly a Type 1 monocyte or T-cell response, which will be further investigated in regard to disease propagation.
To cite this abstract in AMA style:
Mirizio E, Mandal R, Yan Q, Horne W, Schollaert-Fitch K, Torok KS. Genetic Signatures from RNA Sequencing of Pediatric Localized Scleroderma (LS) Skin [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/genetic-signatures-from-rna-sequencing-of-pediatric-localized-scleroderma-ls-skin/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetic-signatures-from-rna-sequencing-of-pediatric-localized-scleroderma-ls-skin/