Session Information
Date: Sunday, October 21, 2018
Title: 3S103 ACR Abstract: Genetics, Genomics & Proteomics: Precision Medicine (916–921)
Session Type: ACR Concurrent Abstract Session
Session Time: 4:30PM-6:00PM
Background/Purpose: To explain the complex regulatory changes associated with RA synovitis and the diversity of responses to targeted therapies, we developed and applied a novel integrative epigenetic profiling method. This method, known as Taiji, focuses on differences between patients with RA in addition to the traditional RA vs. non-RA methodology. Using Taiji, we identified transcription factors (TFs) central to regulatory patterns in fibroblast-like synoviocytes (FLS) that vary between RA patients that could contribute to variable responses to targeted therapies.
Methods: Whole genome ATAC-seq data from 11 RA and 11 osteoarthritis [OA] FLS lines) were evaluated for overlaps between ATAC-seq peaks and known gene promoter regions (4Kb upstream and 1kb downstream from the TSS). Peaks not assigned to promoters were linked with the nearest gene. 745 TFs, with binding motifs curated from the CIS-BP database, had binding sites within 150-bp of ATAC-seq peaks. 22 network topologies were constructed by forming directed edges between any parent node TF and child node gene or child node TF. Via the integration of our whole genome RNA-seq expression data, edge weights were assigned according to the pooled ATAC-seq peak intensity and the expression of the TF parent node. For each sample, the Personalized PageRank (PPR) algorithm was run to measure the global influence of each node and variability between RA patients.
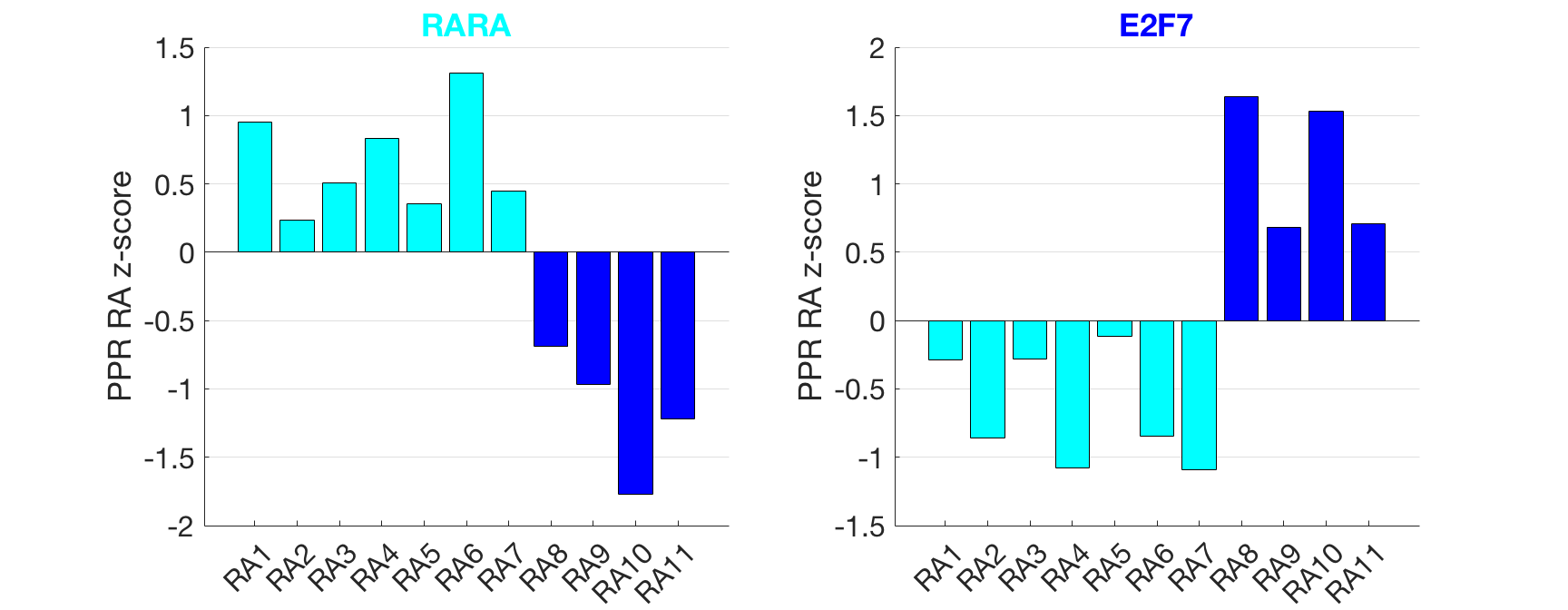
Results: Analysis focused on differences between RA patients. We first clustered the patients using the log2 transcripts per million expression for all genes resulting in two clusters of 7 and 4 RA patients. The mean PPR z-score was calculated for all TFs from each cluster of patients and the absolute difference between the two clusters (DPPR) was calculated. Analysis of the PPR z-score of TF regulatees was conducted to gain statistical power. Ranking TFs by DPPR, the top 200 were intersected with the 310 TFs that have significantly different regulatee PPR z-score values (p<0.003) yielding 80 RA patient-specific TFs that distinguished patients from each other (see Figure showing inter-patient differences in the clusters). High DPPR TFs identified in the two RA clusters included the retinoic acid receptor alpha (RARA) and E2F7 (p=0.003 and p=0.010). RARs play a role in the immunomodulation of synovial inflammation in RA. The E2F TF family member E2F7, plays a role in angiogenesis.
Conclusion: Our novel epigenetic profiling approach defines patient-specific TFs that could contribute to RA patient-to-patient differences. This unique computational approach helps elucidate the mechanism of differential responses to highly targeted agents in RA. Key transcription factors, including genes such as RARA and E2F7, emerge as potential patient-specific TF targets from this in silico method to individualize treatment.
To cite this abstract in AMA style:
Ainsworth R, Zhang K, Zheng L, Firestein GS, Wang W. Rheumatoid Arthritis Patient-Specific Therapeutic Target Identification Using Integrative Epigenetic Profiling [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/rheumatoid-arthritis-patient-specific-therapeutic-target-identification-using-integrative-epigenetic-profiling/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/rheumatoid-arthritis-patient-specific-therapeutic-target-identification-using-integrative-epigenetic-profiling/