Session Information
Session Type: ACR Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose : We investigated 194 individuals with SADs (38 primary Sjögren’s syndrome (pSS), 47 rheumatoid arthritis (RA), 46 systemic lupus erythematosus (SLE), 42 systemic sclerosis (SSc) and 21 undifferentiated connective tissue disease (UCTD) patients) and 53 healthy controls (HCs) to determine whether a fine flow cytometry analysis of T and B cell distribution in whole blood could cluster individuals according to disease diagnosis.
Methods: Two flow cytometry panels were designed. The first panel was dedicated to T cells and combined CD57, CD45RA, CD62L, CD27, CD38, CD3, CD4 and CD8 mAbs. The second panel was dedicated to B cells and combined IgD, TACI, CD27, CD5, CD38, CD19 and CD24 mAbs. A classical manual gating strategy and the Flow-clustering without K (FLOCK) investigation, a density-based clustering approach to algorithmically identify relevant cell populations from multiple samples in an unbiased fashion, were used.
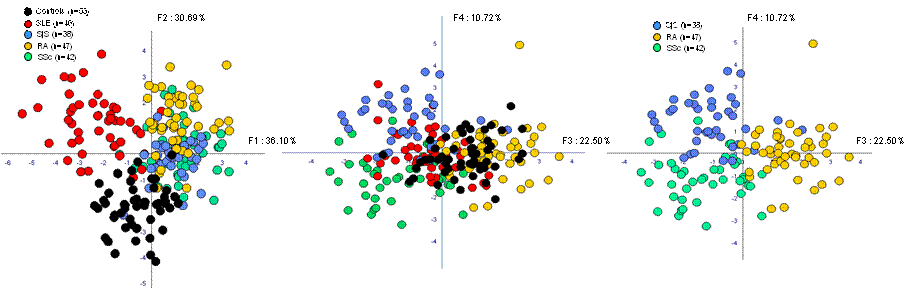
Results: The manual gating strategy allows the identification of 17 distinct lymphocyte subsets. The prediction of the different SADs was determined by discriminant function analysis (DFA). No clustering was found. The FLOCK exploration of the merged HCs identifies 85 distinct subsets of lymphocytes used as reference when compared to SADs . The DFA analysis clearly clusters the HCs and the patients according to each SAD (see figure below).
When compared to HCs, the pSS signature was discriminated by an increase in IgDhiCD24hiCD38hiCD27–TACI–CD5hi transitional B cells, and an increase of CD45RA+CD27–CD62Llo/-CD57hi effector CD8+ T cells.
The SLE signature was discriminated by an increase in IgD–CD24loCD38–CD27–TACI+CD5– memory like B cells, an increase in CD45RA–CD62L+CD38hi activated central memory CD4+ T cells.
The RA signature was discriminated by an increase in IgDhiCD24loCD38–CD27–TACI+CD5– unactivated mature naïve B cells and a decrease in CD45RA+CD62L+CD38hi naïve CD8+ T cells.
The SSc signature was discriminated by a decrease in CD45RA+CD62L+CD38hi naïve CD8+ T cells.
Interestingly, patients with UCTD were distributed among the different clusters (28% with HC, 29% with SLE, 29% with SSc, 9% with RA and 5% with pSS clusters).
Conclusion: A fine bioinformatical flow cytometry analysis of T and B cell subsets clusterizes patients and HCs suggesting that each SAD can be associated with abnormal specific phenotypical distributions that could be helpful in the diagnosis.
This work has received support from the EU/EFPIA Innovative Medicines Initiative Joint Undertaking PRECISESADS grant n° 115565.www.precisesads.eu
To cite this abstract in AMA style:
Simon Q, Rouvière B, Martin T, Le Lann L, Saraux A, Devauchelle-Pensec V, Marañón C, Varela Hernández N, Dufour A, Chizzolini C, de Langhe E, Barbarroja N, Lopez-Pedrera C, Gerl V, Degroof A, Ducreux J, Trombetta E, Li T, Alarcón-Riquelme M, Jamin C, Pers JO. A Fine Bioinformatical Analysis of Lymphocyte Distribution Predicts the Diagnosis of Systemic Autoimmune Diseases [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 10). https://acrabstracts.org/abstract/a-fine-bioinformatical-analysis-of-lymphocyte-distribution-predicts-the-diagnosis-of-systemic-autoimmune-diseases/. Accessed .« Back to 2017 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-fine-bioinformatical-analysis-of-lymphocyte-distribution-predicts-the-diagnosis-of-systemic-autoimmune-diseases/