Session Information
Session Type: Abstract Submissions
Session Time: 5:30PM-7:00PM
Background/Purpose: The goal of this project is the identification of informative synovial biomarkers to predict which children with oligoarticular juvenile idiopathic arthritis (JIA) will have a persistent course, with no more than 4 involved joints, vs those who will have an extended course, with a cumulative total of ≥5 affected joints after the first 6 months of disease.
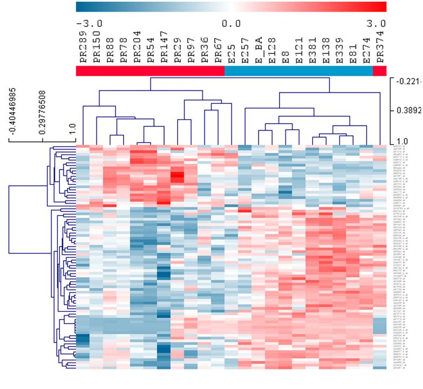
Methods: As part of a separate ongoing IRB approved protocol, remnant synovial fluid was obtained from patients undergoing medically indicated arthrocenteses. All patients satisfied ACR classification criteria for JIA. Using our clinical database, JIA samples were separated into two groups: (1) oligoarticular JIA with persistent course (PR), (2) oligoarticular JIA with extended course (E). All samples were from steroid-na•ve joints and most samples from E were obtained prior to extension. Primary cultures of fibroblast-like synoviocytes (FLS) were established for each subject. RNA from cultured passage 3-6 FLS were isolated, amplified and hybridized to Affymetrix Human GeneChips using the Affymetrix protocol. Expression values were determined with GC-RMA. Global gene expression of FLS from 12 PR and 11 E samples were obtained. Data was filtered for log2 expression >4 in all samples of either E or PR, then for absolute value of 1.5-fold change. Bioconductor package Linear Models for Microarray Analysis (LIMMA) revealed 83 probesets with statistically significant differential expression between E vs PR FLS (7% false discovery rate), shown in heatmap.
Results: Hierarchical clustering of the 83 probesets revealed samples from the different courses cluster together, with most of the PR to the left of the heatmap. Importantly, the all of the E were taken from the very first sample available, which preceded extension in the majority of patients, highlighting that there are detectible differences in the gene expression of the FLS early in the course in the patients whose disease is destined to extend. Of these 83 probesets, 9 corresponded to genes with secreted proteins. We performed mathematical modeling with linear discriminant analysis (LDA) on these 9 genes to reveal 6 genes (KLHL13, MAMLD1, ANKRD44, CD14, HSPBAP1, and MBP) which could correctly predict group, E or PR, 100% of the time using leave-one-out cross validation. ELISA was used to confirm expression of these secreted proteins in synovial fluids.
Conclusion: We were able to demonstrate differential gene expression in FLS from JIA patients who remained PR vs those who were destined to extend, demonstrating detectible difference early in disease which may be useful for prediction. The differentially expressed genes, especially for secreted proteins, provide a starting point for development of biomarkers to distinguish between PR and E JIA using aspirated synovial fluid.
To cite this abstract in AMA style:
Brescia A, Simonds M, McCahan SM, Bunnell T, Sullivan KE, Rosé CD. Linear Discriminant Analysis of Cultured Fibroblast-like Synoviocytes Identifies 6 Candidate Genes Which Predict Extended Course in Juvenile Idiopathic Arthritis [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 4). https://acrabstracts.org/abstract/linear-discriminant-analysis-of-cultured-fibroblast-like-synoviocytes-identifies-6-candidate-genes-which-predict-extended-course-in-juvenile-idiopathic-arthritis-2/. Accessed .« Back to 2017 Pediatric Rheumatology Symposium
ACR Meeting Abstracts - https://acrabstracts.org/abstract/linear-discriminant-analysis-of-cultured-fibroblast-like-synoviocytes-identifies-6-candidate-genes-which-predict-extended-course-in-juvenile-idiopathic-arthritis-2/