Session Information
Session Type: ACR Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: MicroRNAs (miRNAs) are a group of small, noncoding RNAs that post-transcriptionally regulate gene expression. miRNAs play important roles in chondrocyte function and in the development of osteoarthritis. We characterized the dynamic repertoire of the chondrocyte miRNAs and isomiRs by deep sequencing analysis in chondrocytes isolated from normal subjects as well as OA patients in response to the treatment with inflammatory cytokine IL-1b. We also tested the role of TUT7/ZCCHC6 in non-template addition of nucleotides at the ends of specific miRNAs in human chondrocytes.
Methods: Joint cartilage samples were obtained from normal subjects at autopsy and OA patients who underwent joint arthroplasty at the Crystal Clinic, Akron, OH. Primary human chondrocytes were prepared by sequential digestion of the cartilage pieces by pronase (1 mg/ml) for 2 hrs and collagenase A (1 mg/ml) for 16 hrs. Cells were serum starved overnight followed by treatment with IL-1b (2 ng/ml) for different durations (2 hrs, 12 hrs and 24 hrs). Expression of TUT7/ZCCHC6 was knocked down using ON-TARGETplus SMARTpool (GE Dharmacon). Total RNA was prepared using miRNeasy kit (Qiagen) and libraries for sequencing were prepared using True seq small RNA library preparation kit (Illumina). Next generation sequencing was performed on Illumina MiSeq sequencer. Data were analyzed using CLC genomics suite version 8 (Qiagen).
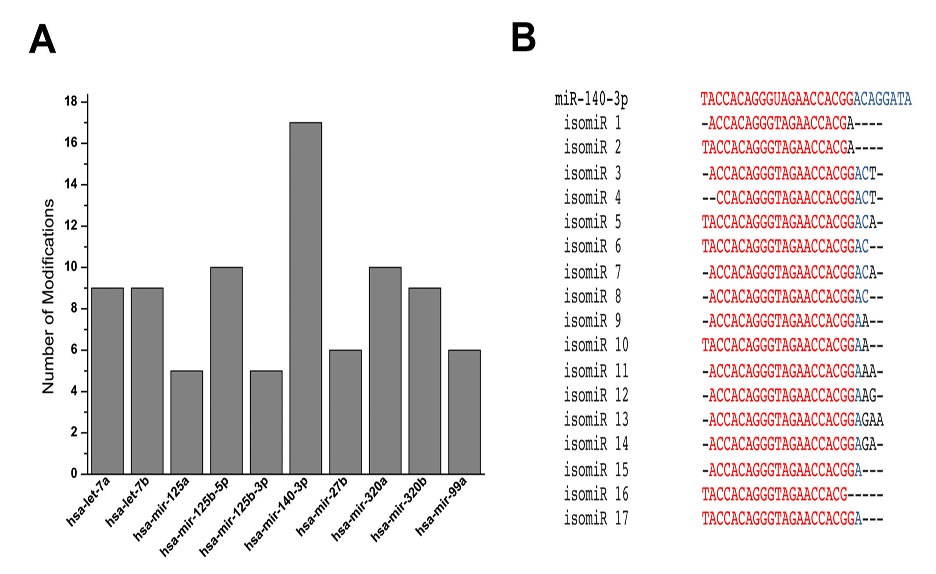
Results: There was a modest difference in the expression of miRNAs in normal and OA chondrocytes in the presence or absence of IL-1b. We found a number of miRNAs that showed a wide range of sequence modifications including nucleotide additions and deletions at 5’ and 3’ ends; and nucleotide substitutions. Some modifications resulted in seed shifts. Interestingly, miR-140-3p, an important miRNA involved in chondrocyte function, was found to have the highest number of modifications (Figure 1). Many of these modifications have been shown to impart a change in the miRNAs’ ability to regulate gene expression. Knockdown of TUT7/ZCCHC6 resulted in a significant change in the isomiR profile in human chondrocytes demonstrating that TUT7/ZCCHC6 contributes to the miRNA complexity in human chondrocytes.
Conclusion: Together, these results reveal a complex repertoire of miRNAs and isomiRs in primary human chondrocytes and their dynamic changes in response to IL-1b treatment. Here we also show the role of TUT7/ZCCHC6 in generating the miRNA diversity in human chondrocytes. These findings will help us better understand miRNA-mediated control of gene expression in the pathogenesis of OA. This work was supported in part by USPHS/NIH grants (RO1 AT007373, RO1 AT005520, RO1 AR067056, R21 AR064890) and funds from North East Ohio Medical University to TMH.
Figure 1. A. Number of modifications in 10 most modified miRNAs with ³100 reads. B. Individual modifications with ³100 read counts in miR-140-3p aligned with the archetype miRBase sequence.
To cite this abstract in AMA style:
Haseeb A, Shahidul Makki M, Ansari M, Piontkivska H, Haqqi TM. Terminal Uridylyl Transferase ZCCHC6-Dependent Generation of miRNA Diversity in Primary Human Chondrocytes [abstract]. Arthritis Rheumatol. 2016; 68 (suppl 10). https://acrabstracts.org/abstract/terminal-uridylyl-transferase-zcchc6-dependent-generation-of-mirna-diversity-in-primary-human-chondrocytes/. Accessed .« Back to 2016 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/terminal-uridylyl-transferase-zcchc6-dependent-generation-of-mirna-diversity-in-primary-human-chondrocytes/