Session Information
Date: Monday, November 9, 2015
Title: Cytokines, Mediators, Cell-cell Adhesion, Cell Trafficking and Angiogenesis II
Session Type: ACR Concurrent Abstract Session
Session Time: 2:30PM-4:00PM
Background/Purpose: We investigated mechanisms driving pathogenic behavior of synovial fibroblasts (FLS) in rheumatoid arthritis (RA).
Methods: FLS from RA patients (1987 classification criteria) were extracted. Expression of 249 inflammation-related genes was measured by nanoString technology. Active transcription was evaluated by primary transcripts. Chromatin accessibility was assessed by FAIRE assay. mRNA decay rates, after blocking transcription with actinomycin D (ActD), were calculated in a semi-log plot. The role of NF-kB and MAPKs was tested with inhibitors iIKK, SB202190, SP600125, U0126.
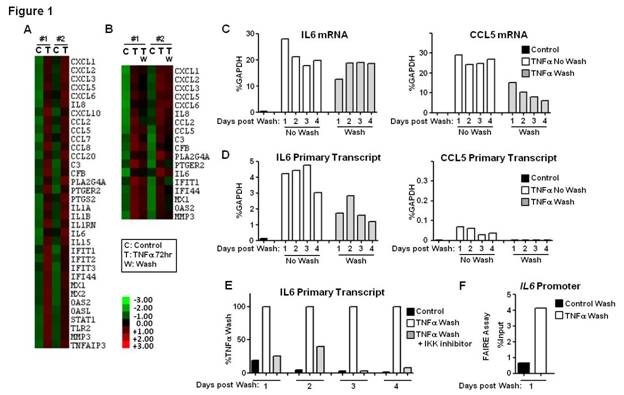
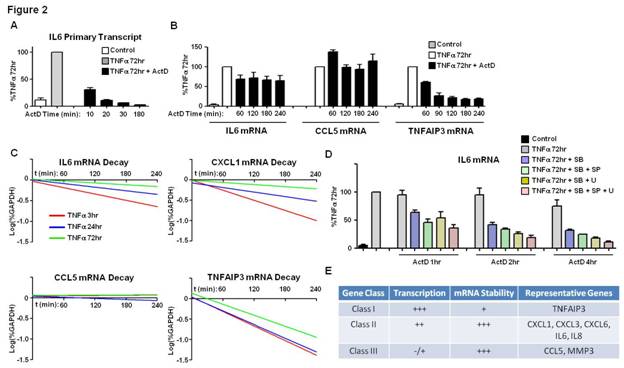
Results: Using strict criteria (expression level >100 at 72h of TNF-stimulation and >3-fold induction compared to control), a cluster of 34 genes displayed sustained induction (2 independent experiments) (Figure 1A). 18/34 genes retained high mRNA expression levels despite removal (washing) and blockade (infliximab) of TNFa (Figure 1B-C). In unwashed conditions, active transcription at TNF 72h was variably present depending on the gene. High levels of active transcription were maintained in genes such as IL6, IL8, CXCL1, CXCL2, CXCL3, CXCL6 and TNFAIP3, even after removal of TNF (Figure 1D and not shown). Continuous transcription was NF-kB dependent (Figure 1E). Sustained IL6 promoter chromatin remodeling was retained for days post-wash (Figure 1F and not shown). Highly expressed genes, such as CCL5 and MMP3, displayed minimal ongoing transcription (Figure 1D and not shown). Prolonged (4-8h) inhibition of transcription with ActD at TNF 72h (Figure 2A) significantly decreased TNFAIP3 mRNA (Figure 2B). Expression of other genes, such as IL6, IL8, CXCL1, CXCL3, CXCL6, CCL5 and MMP3, was minimally affected by ActD (Figure 2B and not shown), suggesting increased mRNA stability. Decay rates of mRNA for a group of TNF-inducible genes, such as IL6 and CXCL1, gradually decreased with time (Figure 2C), indicating TNF-inducible mRNA stabilization effect, which was MAPK-dependent (Figure 2D). Decay rates of mRNA of other genes were not dynamically regulated by TNF, and were either persistently stable (e.g CCL5) or unstable (e.g TNFAIP3) (Figure 2C).
Conclusion: We have identified a sustained inflammation-related gene expression program in TNF-stimulated RA FLS. Within this program there are different classes of genes based on the relative contribution of transcription and mRNA stability (Figure 2E). TNF drives MAPK-dependent stabilization of certain transcripts and persistent chromatin remodeling in genes such as IL6, indicating TNF-induced memory effect in FLS.
To cite this abstract in AMA style:
Loupasakis K, Sohn C, Ivashkiv LB, Kalliolias GD. TNF Confers Pathogenic Memory in Synovial Fibroblasts Via Chromatin Remodeling, NF-Kb-Dependent Transcription and MAPK-Mediated mRNA Stabilization [abstract]. Arthritis Rheumatol. 2015; 67 (suppl 10). https://acrabstracts.org/abstract/tnf-confers-pathogenic-memory-in-synovial-fibroblasts-via-chromatin-remodeling-nf-kb-dependent-transcription-and-mapk-mediated-mrna-stabilization/. Accessed .« Back to 2015 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/tnf-confers-pathogenic-memory-in-synovial-fibroblasts-via-chromatin-remodeling-nf-kb-dependent-transcription-and-mapk-mediated-mrna-stabilization/