Session Information
Date: Tuesday, November 15, 2016
Title: Systemic Lupus Erythematosus – Clinical Aspects and Treatment IV: Biomarkers
Session Type: ACR Concurrent Abstract Session
Session Time: 4:30PM-6:00PM
Background/Purpose: SLE patients can experience chronically active disease, remissions and flares, or long periods of quiescence. Accurately assessing disease activity is crucial for prescribing appropriate drug regimens, evaluating treatment outcomes, and detecting early onset of flares; however, there is currently no “gold-standard” biomarker for this determination. Though not in routine clinical use, the SLEDAI scoring system is often applied in clinical trials to evaluate the efficacy of new drugs. Scores
Methods: Sera from 500 SLE patients meeting the ACR classification criteria at diagnosis were evaluated by IMS at different times post-diagnosis. SLEDAI scores ranged from 0 (n=150) to 27. Patients with scores 2-27 (n=350) were selected to sample the range as uniformly as possible (median = 8; IQR 4-10). IMS assays were used to measure serum antibodies bound to a microarray of ~126,000 unique peptides designed to broadly sample chemical space, thereby providing a library of diverse epitope mimetics for antibodies to selectively bind. Peptide features with binding intensities that were significantly different between active and inactive SLE were identified by t-test. Support vector machine classifiers and linear regression models were trained and tested by 100 iterations of 5-fold cross validation.
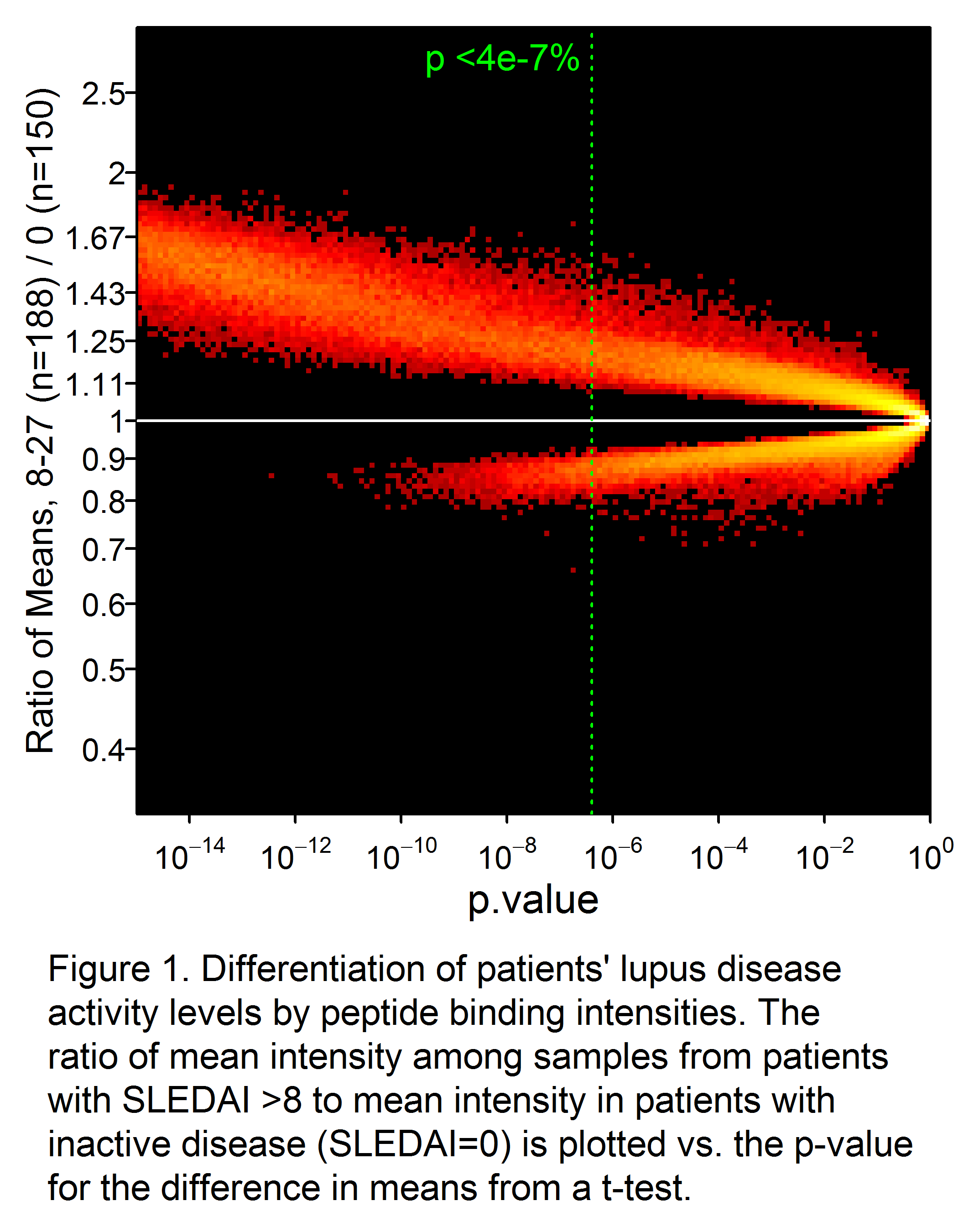
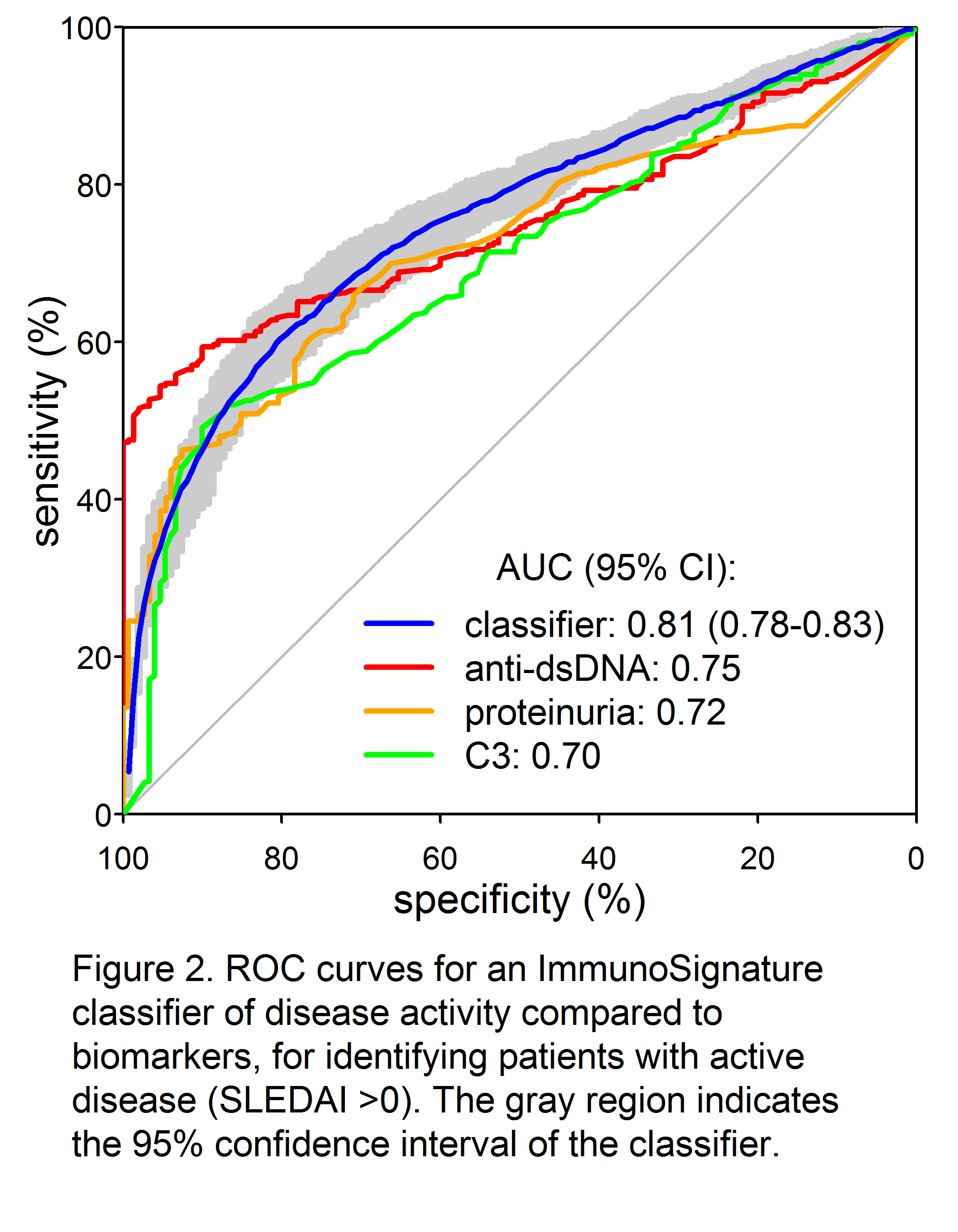
Results: A total of 34,147 peptides were identified that significantly (p<0.05 after Bonferroni correction) differentiated active from inactive SLE (Fig 1). Receiver-operator characteristic (ROC) curves demonstrated significantly superior classification of the samples as inactive or active by an IMS classifier model, compared to standard biomarkers of disease activity such as C4 (not shown), C3, anti-dsDNA, and proteinuria (Fig. 2). Good discrimination was achieved by training on SLEDAI >8 vs. 0, with performance further enhanced when applied to extreme contrasts. For example, a classifier of SLEDAI >15 vs. 0 had an AUC of 0.90 (95% CI 0.88 – 0.92). Correlations of a linear IMS (r=0.48), C3 (r= -0.41) and anti-dsDNA (r=0.36) to SLEDAI were also determined.
Conclusion: A single serologic test, using specific binding patterns of serum antibodies on a highly complex and diverse mimetic peptide array, can molecularly determine SLE disease activity and improve upon current methods of evaluating and monitoring patients.
To cite this abstract in AMA style:
Putterman C, Rowe M, Legutki JB, Tarasow TM, Sykes K. A Simple Test for Assessing and Monitoring SLE Disease Activity Status [abstract]. Arthritis Rheumatol. 2016; 68 (suppl 10). https://acrabstracts.org/abstract/a-simple-test-for-assessing-and-monitoring-sle-disease-activity-status/. Accessed .« Back to 2016 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-simple-test-for-assessing-and-monitoring-sle-disease-activity-status/