Session Information
Date: Wednesday, November 8, 2017
Title: Systemic Sclerosis, Fibrosing Syndromes and Raynaud's – Pathogenesis, Animal Models and Genetics II
Session Type: ACR Concurrent Abstract Session
Session Time: 9:00AM-10:30AM
Background/Purpose: Systemic sclerosis (SSc) is a multisystem disease that has a higher prevalence in African Americans (AA), with a more severe phenotype, internal organ involvement, and increased mortality. Genetic risk factors contributing to the AA SSc phenotype are largely unknown.
Methods: The Genome Research in African American Scleroderma Patients consortium (GRASP) was established to recruit AA patients to investigate the genetic basis of SSc. The Illumina MEGA array was used to genotype 934 patients and 946 controls to conduct the first genome-wide association study (GWAS) of SSc in AA. A total of 1.7 million variants were genotyped and after quality control filtering 1.4 million variants remained and were imputed into the 1000 Genomes Phase 3 v5 reference panel. We utilized HiChIP, a protein-centric chromatin conformation method, to elucidate the role of these variants.
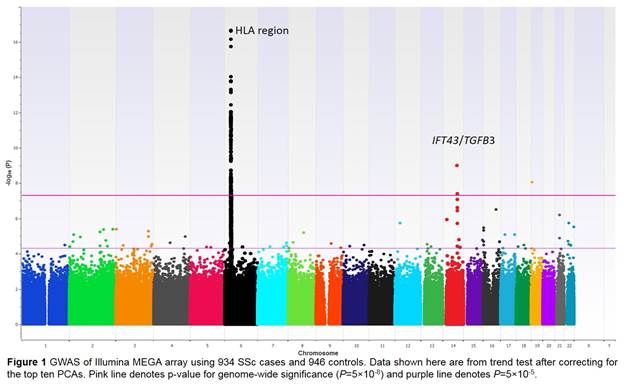
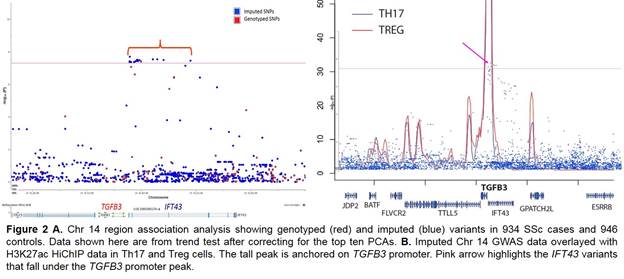
Results: Admixture and principal component analysis (PCA) confirmed that the patients and controls were well-matched. λGC was 1.03, suggesting no major population stratification. The MHC region demonstrated the strongest association after accounting for the top 10 PCAs (Figure 1). The rs35915063 was the top scoring variant in the HLA-DQB1 gene P=2.2 x 10-17, OR=1.96 (95% CI 1.7-2.3). Stepwise conditional regression analysis using an additive model revealed the DQA1 gene variant rs9271620 (P=1.01 x 10-8) and the DPB1 gene variant rs2071354 (P=5.8 x 10-8) as independent risk variants. eQTL databases revealed a reduction in DQB1, DQA1, and DRB1 expression and an increase in DPB1 and DQA2 expression based on the associated allele of these three variants. The rs59063398 variant showed the strongest non-MHC association with P=2.0 x 10-8, OR=0.47 (95% CI 0.4-0.6) (Figure 2A). This variant is part of an evolutionarily conserved haploblock in the IFT43 gene region that is African ancestry specific. Overlaying our GWAS data on the H3K27ac HiChIP data in Th17 and Treg cells highlighted IFT43 intronic variants falling under the TGFB3 promoter peak, suggesting potential alteration of TGFB3 promoter activity and in turn regulation of TGFB3 expression (Figure 2B).
Conclusion: This first GWAS of SSc in AA confirms MHC as a susceptibility locus and identifies a novel (TGFB3), non-MHC locus, as a SSc susceptibility gene. IFT43/TGFB3 region variants are non-polymorphic in Europeans, suggesting an explanation for the increased prevalence and severity of SSc in AA. TGFB3 has a role in fibrosis and in development of effector Th17 cells, raising the possibility of therapies targeting these pathways.
To cite this abstract in AMA style:
Gourh P, Remmers EF, Satpathy A, Boyden S, Morgan ND, Shah AA, Adeyemo A, Bentley A, Carns MA, Chandrasekharappa SC, Chung L, Criswell LA, Derk CT, Domsic RT, Doumatey A, Gladue H, Goldberg A, Gordon JK, Hsu V, Jan R, Khanna D, Mayes MD, Medsger TA Jr., Mumbach M, Ramos PS, Trojanowski M, Saketkoo LA, Schiopu E, Shanmugam VK, Shriner D, Silver RM, Steen VD, Valenzuela A, Varga J, Chang H, Rotimi C, Wigley FM, Boin F, Kastner DL. Transforming Growth Factor Beta 3 (TGFB3) – a Novel Systemic Sclerosis Susceptibility Locus Involved in Fibrosis and Th17 Cell Development Identified By Genome-Wide Association Study in African Americans from the Genome Research in African American Scleroderma Patients Consortium [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 10). https://acrabstracts.org/abstract/transforming-growth-factor-beta-3-tgfb3-a-novel-systemic-sclerosis-susceptibility-locus-involved-in-fibrosis-and-th17-cell-development-identified-by-genome-wide-association-study-in-african-americ/. Accessed .« Back to 2017 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/transforming-growth-factor-beta-3-tgfb3-a-novel-systemic-sclerosis-susceptibility-locus-involved-in-fibrosis-and-th17-cell-development-identified-by-genome-wide-association-study-in-african-americ/