Session Information
Date: Monday, November 13, 2023
Title: Abstracts: SLE – Diagnosis, Manifestations, & Outcomes II: Omics
Session Type: Abstract Session
Session Time: 4:00PM-5:30PM
Background/Purpose: Iberdomide is a high affinity cereblon ligand that promotes ubiquitylation and proteasomal degradation of Ikaros (IKZF1) and Aiolos (IKZF3) transcription factors and, thereby altering specific aspects of immune responsiveness. Iberdomide has been shown to be efficacious in a randomized controlled trial in patients with generalized SLE (NCT03161483) and to be specifically effective in patients with high baseline expression of the interferon gene signature (IGS)1,2. The current analysis sought to identify the profile of gene expression abnormalities in SLE patients responsive to iberdomide and the impact of the agent on gene expression abnormalities.
Methods: Baseline whole blood samples from 276 female SLE patients from the phase 2b iberdomide trial were utilized for this analysis. These patients had a >/= 6 month history of SLE and disease activity determined by SLEDAI-2K >/=6. Patients were randomized to placebo, or one of three doses of iberdomide (0.15, 0.3 or 0.45 mg once daily). Clinical response was determined by the SLE Responder Index 4 (SRI-4) at 24 weeks. RNAseq was performed and analyzed by Gene Set Variation Analysis (GSVA) using 32 informative gene modules and K-means clustering.
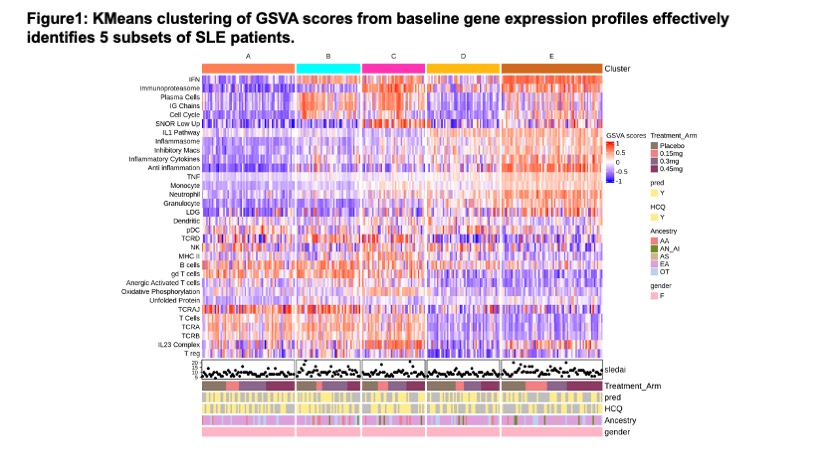
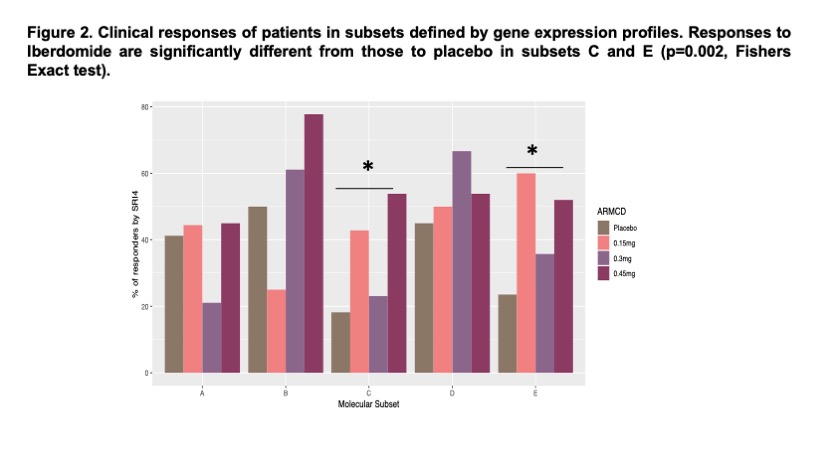
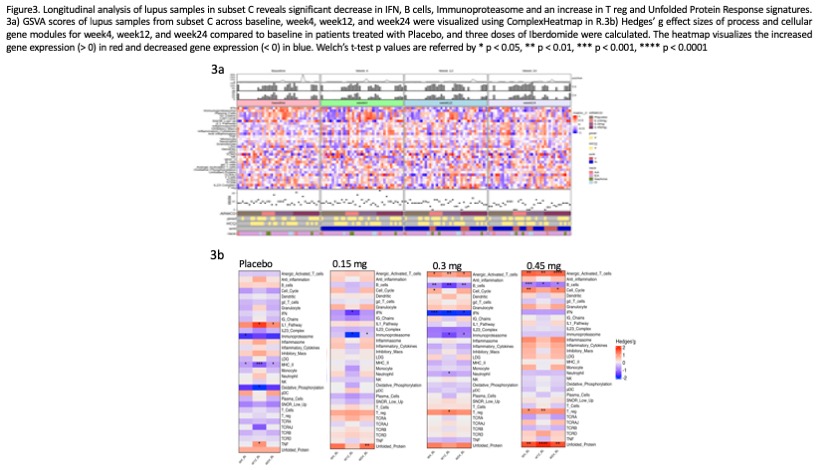
Results: Whole blood K-means clustering of the GSVA scores yielded 5 subsets of patients (Figure 1). Subset A had the fewest molecular abnormalities, whereas Subset E had the most disturbances in immune function, including enrichments in the IGS, immunoproteasome, IL-1/ inflammasome pathway, and neutrophil/granulocyte genes and lymphopenia. Clusters B-D had intermediate degrees of abnormal enrichment in specific gene modules. Cluster C had high IGS, immunoproteasome, plasma cells/Ig chains, and IL-23 complex genes, but no lymphopenia. No differences were noted between the subsets with regard to steroid or hydroxychloroquine use, and differed only modestly in disease activity as measured by SLEDAI-2K, anti-DNA and Compelemnt C3and C4. Significant clinical responses to iberdomide were confined to subsets C and E (Figure 2). Effect sizes of responses in these groups ranged between 20-30%. Other subsets had higher placebo responses and no additional response to iberdomide. Treatment with iberdomide resulted in significant decreases in the B cell, plasma cell and interferon signatures and increases in the Treg signature (Figure 3).
Conclusion: K-means clustering of GSVA scores from baseline samples of the iberdomide trial successfully clustered patients into subsets that exhibited differences in response to iberdomide treatment, with the greatest responses observed in patients with the highest IGS, immunoproteasome, plasma cell, inflammasome, and IL-23 pathways. Treatment with iberdomide altered gene expression profiles in a manner consistent with the known action of the agent. Gene expression based subsetting may be useful to enrich trials for responsive patients and monitor the impact of therapy.
To cite this abstract in AMA style:
Bachali P, Korish S, Hu Y, Schafer P, Grammer A, Lipsky P. Transcriptomic Analysis of the Impact of Iberdomide on Patients with SLE [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/transcriptomic-analysis-of-the-impact-of-iberdomide-on-patients-with-sle/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/transcriptomic-analysis-of-the-impact-of-iberdomide-on-patients-with-sle/