Session Information
Date: Wednesday, November 13, 2019
Title: 6W013: SLE – Clinical VI: Epidemiology, Diagnosis, & Outcomes (2882–2887)
Session Type: ACR Abstract Session
Session Time: 9:00AM-10:30AM
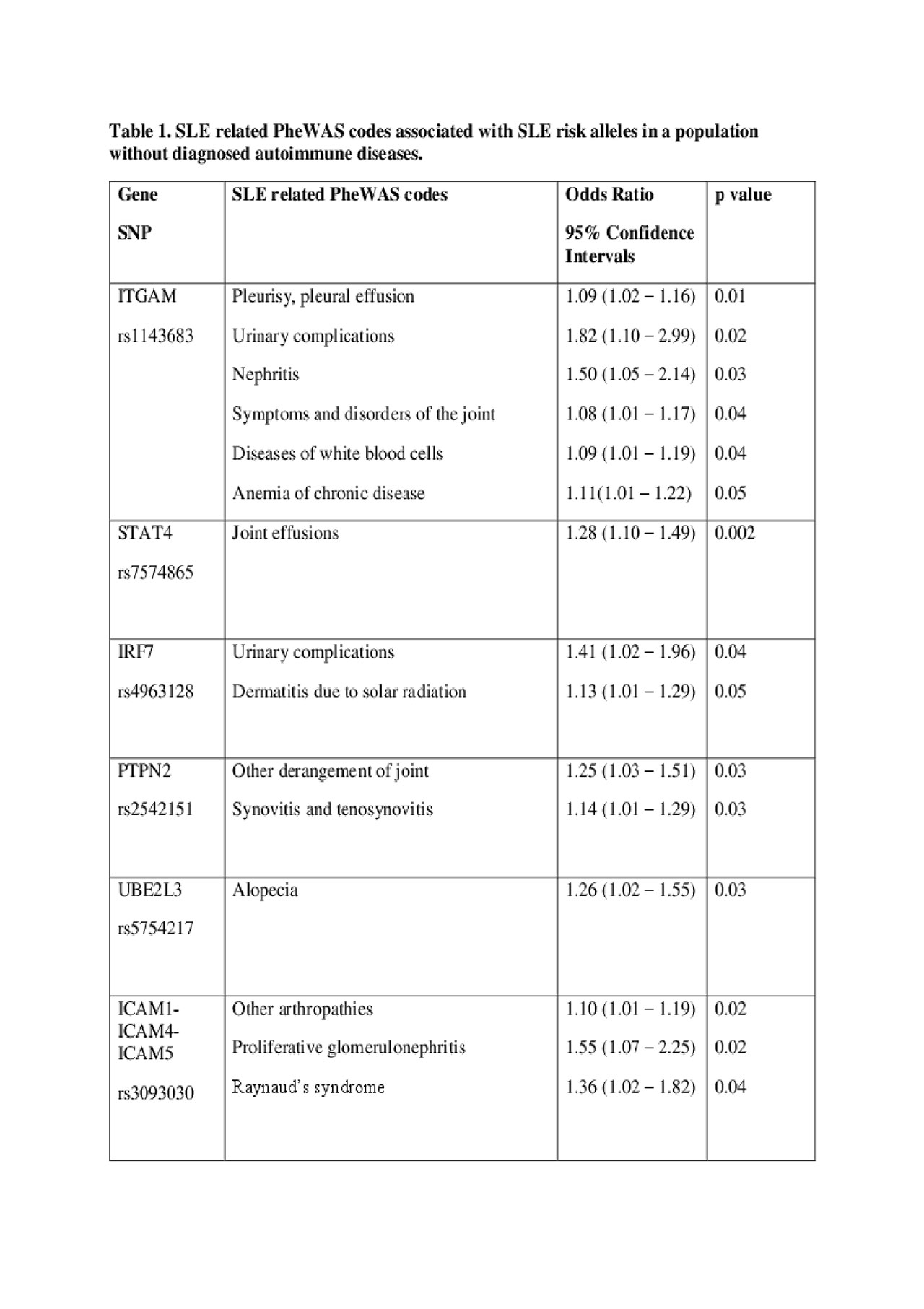
Background/Purpose: Systemic lupus erythematosus (SLE) is a heterogeneous disease with > 100 known risk alleles. Some of these risk alleles associate with ACR SLE criteria such as renal disease. We examined if nine of these SLE risk alleles would identify autoimmune features in a population without diagnosed autoimmune diseases.
Methods: We investigated subjects from BioVU, a databank that links DNA extracted from routine, clinical blood samples to a de-identified electronic health record (EHR) with several decades of clinical data. BioVU contains 250,000 subjects, over 80,000 of which have been genotyped on Illumina MEGA chip. We selected individuals who had available data for 9 SLE risk single nucleotide polymorphisms (SNPs) (ITGAM rs1143683 and 1143679, STAT4 rs7574865, IRF5 rs10488631, IRF7 rs4963128, TFAF3IP2 rs13190932, PTPN2 rs2542151, UBE2L3 rs5754217, ICAM1-ICAM4-ICAM5 rs3093030), which have been associated with ACR SLE criteria. Subjects did not have ICD9 or ICD10 billing codes for autoimmune diseases including SLE, rheumatoid arthritis, Sjogren’s syndrome, systemic sclerosis, or myositis. We compared subjects with or without these nine SLE risk SNPs using phenome-wide association studies (PheWAS). PheWAS is a technique that scans across EHR billing codes. Subjects with a SLE risk SNP and multiple codes corresponding to SLE ACR criteria were chart reviewed to determine if they had an undiagnosed autoimmune disease and assessed for other comorbidities.
Results: We identified 56,496 Caucasian subjects without billing codes for autoimmune diseases that had available genetic data for the 9 SLE risk SNPs. Of these subjects, 56% were female with a mean age of 53 years. Compared to subjects without the SNPs, subjects with the risk SNPs were more likely to have codes related to ACR SLE criteria including nephritis, arthritis, serositis, and hematologic disorders (Table 1, Figure 1). Subjects with the ITGAM rs1143683 SNP were more likely to have codes related to nephritis (p = 0.03), arthritis (p = 0.04), and diseases of white blood cells (p = 0.04). Renal codes were also associated with IRF7 rs4963128 (p = 0.04) and ICAM1-ICAM4-ICAM5 rs3093030 (p = 0.02). On chart review of the 50 subjects with the greatest number of codes related to ACR SLE criteria, 2 subjects fit SLICC SLE criteria but were not formally diagnosed with SLE. Both of these subjects had renal biopsies suggestive of SLE nephritis. An additional two subjects had incomplete SLE and 1 subject ANCA-associated vasculitis, 1 amyloidosis, and 2 Henoch-Schonlein purpura. Notably, 45 (90%) of the subjects had renal disease with 19 requiring dialysis or a renal transplant.
Conclusion: Carriage of SLE risk alleles confers risk of SLE features in undiagnosed subjects. Some of these subjects may be pre-SLE or undiagnosed SLE cases or other autoimmune disease cases. The combination of SLE risk alleles plus early features of the disease may identify a population who are at high risk for developing clinical SLE who would be candidates for early treatment. Subjects with SLE risk alleles had a significant burden of renal disease. SLE risk alleles may share common pathways for renal disease and act as accelerants for organ damage.
To cite this abstract in AMA style:
Barnado A, Wheless L, Crofford L, Denny J. Systemic Lupus Erythematous Risk Alleles Drive Autoimmune Features in a Population Without Diagnosed Autoimmune Diseases [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/systemic-lupus-erythematous-risk-alleles-drive-autoimmune-features-in-a-population-without-diagnosed-autoimmune-diseases/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/systemic-lupus-erythematous-risk-alleles-drive-autoimmune-features-in-a-population-without-diagnosed-autoimmune-diseases/