Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
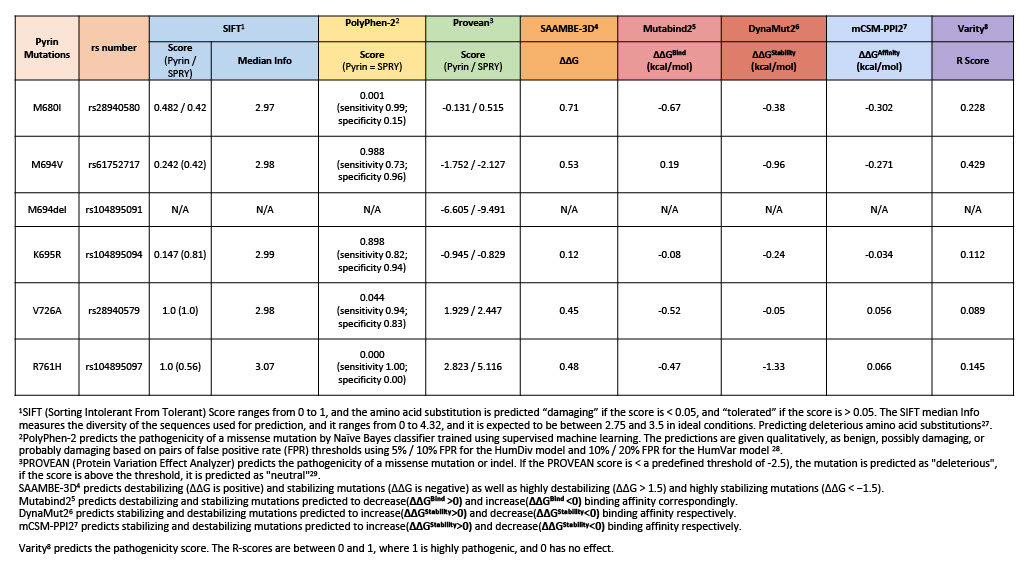
Background/Purpose: Predicting the pathogenicity of amino acid substitutions is crucial for understanding the functional consequences of genetic variations. Several computational methods frequently used so far have been unhelpful in assessing the pathogenicity of mutations in some diseases, such as p.M694V and p.M680I variants in the SPRY domain of pyrin as the main pathogenic variations leading to Familial Mediterranean Fever (FMF) (Table 1). Mutual information (MI) quantifies the degree of information shared between two random variables, which can represent fluctuations in different properties, including biological, chemical, electromagnetic, or mechanical attributes of residues. We herein investigated the role of MI as a measure to quantify the correlation between residue motions or fluctuations as a novel method to predict the pathogenicity of mutations.
Methods: We focused on fluctuations in the positions of residues over time. MI between two residues served as a measure of the statistical dependence or correlation between their respective motions or fluctuations. MI quantified the amount of information shared between the behaviors of the two residues, reflecting how knowing the state or behavior of one residue informed us about the state or behavior of the other residue, and vice versa. We employed extensive molecular dynamics (MD) analyses to investigate the shifts in mutual information profiles between the WT and 5 SPRY domain mutations and 1 deletion (pathogenic mutations M694V, M680I, V726A, and likely pathogenic mutations K695R, R761H; and deletion variant M694del) associated with varying degrees of severity of FMF. We also performed steered MD simulations to investigate the change in the binding affinity of the variants of the SPRY domain to actin. The MI profile shifts were used to quantify the changes upon mutation.
Results: MD simulations of the WT and the mutants revealed that the cavity between two loops of SPRY; one from residue 672 to 680 and the other loop from residue 695 to 697, changed drastically in between mutants, and there was a clear drop in the cavity area of the most penetrant M694V mutation. The M694V mutation exhibited the most substantial negative MI profile shift, followed by the M680I, V726A, and the K695R mutations with a negligible negative shift; and positive shifts were ordered as K695R >M680I >V726A >M694V. The relatively less penetrant V726A was not located in the protein binding cavity, and this replacement resulted in allosteric changes leading to a decrease of MI along a path between A726 and M680, and an increase of the distance between A726 and S675. Deletion of M694, the most severe variant in the SPRY domain, led to extreme loss of MI.
Conclusion: The current work demonstrated the utility of MI profile shifts in assessing the pathogenicity of mutations and the challenges in relating the shifts to disease severity in FMF. The importance of negative MI shifts as indicators of severe effects were emphasized, along with exploring potential compensatory mechanisms indicated by positive MI shifts, which were otherwise random and inconsequential. The current approach is expected to provide insights into the molecular mechanisms underlying disease pathogenesis as well.
To cite this abstract in AMA style:
Hacisuleyman A, Gul A, Erman B. Role of Mutual Information Profile Shifts in Assessing the Pathogenicity of Mutations on Protein Functions: The Case of Pyrin Mutations in Familial Mediterranean Fever [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/role-of-mutual-information-profile-shifts-in-assessing-the-pathogenicity-of-mutations-on-protein-functions-the-case-of-pyrin-mutations-in-familial-mediterranean-fever/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/role-of-mutual-information-profile-shifts-in-assessing-the-pathogenicity-of-mutations-on-protein-functions-the-case-of-pyrin-mutations-in-familial-mediterranean-fever/