Session Information
Session Type: ACR Concurrent Abstract Session
Session Time: 4:30PM-6:00PM
<h1> Background/Purpose: </h1> Immune-mediated
inflammatory disorders (IMIDs) share many genetic risk factors. Pleiotropy may
exist at different levels and most of the underlying mechanisms are still to be
uncovered. GWAS have identified hundreds of risk loci for IMIDs but causative
genes have been identified in only a handful of cases. Recent fine-mapping
efforts indicate that only a minority of risk variants are coding. This
suggests that most risk variants will be regulatory hence affecting disease
risk via eQTL effects.
<h1> Methods: </h1> To aid in
the identification of causative genes for IMIDs, we generated transcriptome
information (HT12 arrays) for six blood cell types (CD4, CD8, CD19, CD14, CD15
and platelets) and intestinal biopsies at three anatomical locations (ileum,
colon, rectum) for 350 healthy Caucasians. The same individuals were genotyped
with SNP arrays interrogating > 700K variants, augmented by imputation from
the 1KG project. To detect cis-eQTL we tested variants within 0.5 megabase
windows centered on the tested probe. The nominal p-value of the best SNP
within a cis-window was Sidak-corrected for the window-specific number of
independent tests. The corresponding best, Sidak-corrected p-values for each
probe were jointly used to estimate their respective false discovery rate. To
identify likely causative genes in GWAS identified risk loci variants and also
better understand pleiotropic effects, we (i) developed a method that
quantifies the correlation between “disease association pattern” (DAP) and
“eQTL association pattern” (EAP) and provides an empirical estimate of its
significance, and (ii) evaluated the effect of fitting known risk variants as
covariates in the eQTL analysis following Nica et al. (2010). We applied both
approaches to celiac disease (CE) and rheumatoid arthritis (RA) and the second
one to type one diabetes (T1D), multiple sclerosis (MS), systemic lupus
erythematosus (SLE), ankylosing spondylitis (AS) and psoriasis (PSO).
<h1> Results: </h1> We detected
> 16000 significant cis-eQTL, with a degree of sharing between cell types
ranging from 38 to 90% highlighting the utility of our multi-tissue panel. GWAS
variants were drivers of ciseQTL effects across the different tissues in 399
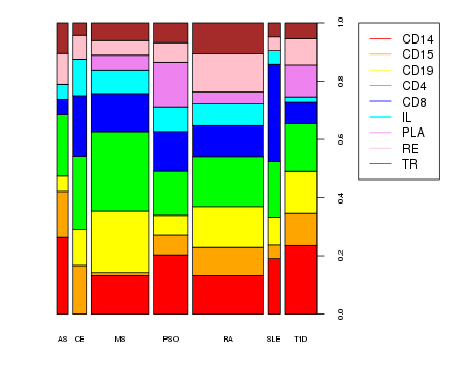
tests (23.6%), mostly in CD4 cells (Figure 1), and pinpointing 64 new
gene-disease associations (3.7%). The number of shared loci and shared eQTL
were highly correlated (rho=0.66).RA and SLE showed the highest degree of sharing.
<h1>Conclusion: </h1> We
identified new potential candidate genes for IMIDs and characterized pleiotropic
effects in terms of sign, magnitude and target tissue, through ciseQTL mapping
in GWAS loci. These findings could shed a light on IMIDs pathogenesis and
co-occurrence. Latest results will be presented.
Figure 1: Tissue Distribution of eQTLs
driven by GWAS loci across IMIDs
Columns’ width is proportional to the
number of eQTLs.
To cite this abstract in AMA style:
Docampo E, Fang M, Dmitrieva J, Théâtre E, Elansary M, Mariman R, Gori AS, Mni M, Crins F, Coppieters W, Louis E, Georges M. Prioritizing Likely Causative Genes in Genome-Wide Association Studies (GWAS) Identified Risk Loci for Immune-Mediated Inflammatory Disorders Using Cell-Type Specific Expression Quantitative Loci (eQTL) Information [abstract]. Arthritis Rheumatol. 2015; 67 (suppl 10). https://acrabstracts.org/abstract/prioritizing-likely-causative-genes-in-genome-wide-association-studies-gwas-identified-risk-loci-for-immune-mediated-inflammatory-disorders-using-cell-type-specific-expression-quantitative-loci-eqt/. Accessed .« Back to 2015 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/prioritizing-likely-causative-genes-in-genome-wide-association-studies-gwas-identified-risk-loci-for-immune-mediated-inflammatory-disorders-using-cell-type-specific-expression-quantitative-loci-eqt/