Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose:
The established association between IL10 and multiple autoimmune diseases including SLE and elevated levels of IL-10 in SLE patients correlating with disease activity led us to fine-map theIL10 family locus (IL10-IL19-IL20-IL24) to evaluate SLE-associated SNPs for their potential functions in multiple ancestries.
Methods:
We genotyped 19 tag SNPs of the 154 kb IL10 family locus in 15533 subjects including European Americans (EA, 3820 cases vs 3412 controls), African Americans (1670 vs 1904), Asians (1252 vs 1249) and Hispanics (1445 vs 781), and imputed an additional 109 SNPs in EA. Each SNP was assessed for association with SLE, and haplotype-based conditional testing was conducted to distinguish independent signals. Transcript and protein levels of IL-10 were measured by real-time PCR and ELISA. EMSA was performed using nuclear lysates of SLE PBLs to assess differential binding to SLE-associated SNP alleles that might regulate IL-10 expression. Elk-1 activation was detected by Western blot, and co-localization of IL-10 and phospho-Elk-1 by flow cytometry.
Results:
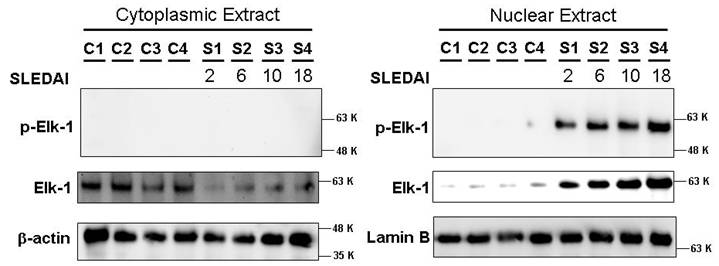
Among 19 genotyped SNPs, the 3’UTR downstream SNP of IL10, rs3024505, exhibited the strongest independent association with SLE susceptibility in EA only (P = 2.67×10-8, OR [95%CI] =1.30 [1.19-1.43]). Lower allelic frequencies of this SNP and lack of other association signals in non-EA led us to focus on EA for imputation, showing association with SLE of 3 additional SNPs: rs3122605 (10kb 5′ upstream), rs3024493 (intron3) and rs3024495 (intron4) tagged by rs3024505 (r2>0.91). SLE patients carrying risk-alleles of these 4 SNPs had higher IL10 expression at mRNA (P=3.8 x 10-6) and protein level (P=4.2 x 10-5) than those carrying non-risk alleles. Only the risk allele of rs3122605 exhibited binding to nuclear extracts from SLE PBLs using EMSA. This risk allele was predicted to preferentially bind to transcription factor Elk-1, and was validated by supershift in the presence of ELK-1 antibodies, suggesting it is the likely causal variant. Upon activation, cytoplasmic Elk-1 is known to be phosphorylated and translocated into the nucleus inducing transcription. Phospho-Elk-1 was detected in nuclear extracts from SLE but not normal PBMCs, and appeared higher in patients with increasing SLEDAI scores (Figure). Co-expression of phospho-Elk-1 and IL-10 in PBLs was elevated in SLE than controls (P = 0.005) and active than inactive patients (P < 0.05).
Conclusion:
We identified GWAS level association (P<5×10-8) of 4 IL10 SNPs with SLE in EA. The risk allele of 5′ upstream rs3122605 preferentially binding to ELK-1 was associated with elevated IL10 levels. Nuclear localization of activated phospho-Elk-1 was elevated in SLE PBLs that also expressed IL-10, especially during active disease. Taken together, the SLE-associated rs3122605 C allele conferred SLE risk by upregulating ELK-1-mediated IL-10 expression.
Disclosure:
D. Sakurai,
None;
J. Zhao,
None;
Y. Deng,
None;
J. A. Kelly,
None;
K. Moser Sivils,
None;
K. M. Kaufman,
None;
E. E. Brown on behalf of PROFILE,
None;
M. E. Alarcón-Riquelme on behalf of BIOLUPUS and GENLES network,
None;
J. B. Harley,
None;
S. C. Bae,
None;
C. O. Jacob,
None;
T. J. Vyse,
None;
T. B. Niewold,
None;
P. M. Gaffney,
None;
J. A. James,
None;
R. P. Kimberly,
None;
G. S. Gilkeson,
None;
D. L. Kamen,
None;
C. D. Langefeld,
None;
D. M. Chang,
None;
Y. W. Song,
None;
W. Chen,
None;
J. M. Grossman,
None;
B. H. Hahn,
None;
B. P. Tsao,
None.
« Back to 2012 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/preferential-binding-to-elk-1-by-sle-associated-il10-risk-allele-up-regulates-il10-expression/