Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Rheumatic diseases often require immunosuppressive treatment, increasing the risk of infection. Clinicians frequently face challenges distinguishing between infection and disease flare in these patients, as both can present with similar symptoms. This study aims to develop a machine learning model to predict the risk of infection in hospitalized patients with rheumatic diseases, using data from the MIMIC-IV clinical database, to aid in clinical decision-making and improve patient outcomes.
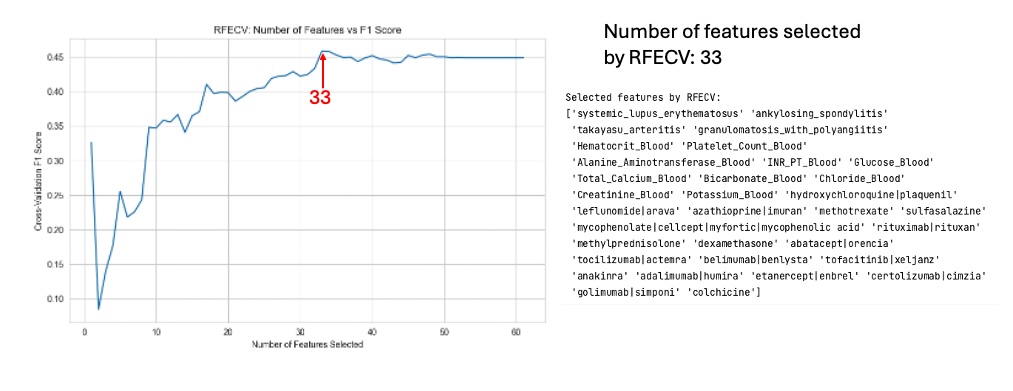
Methods: Data were extracted from the MIMIC-IV v2.2 database, encompassing 431,231 admissions and 299,712 unique patients. Patients with rheumatic disease diagnoses were identified using ICD-9 and ICD-10 codes. Initial laboratory results, vital signs, microbiology culture results, and discharge summaries were collected. A final dataset of 2841 admissions was prepared, with 918 positive and 1923 negative culture results, indicating class imbalance. Feature selection was performed using recursive feature elimination, and models were developed using Logistic Regression, Random Forest, and XGBoost algorithms. Hyperparameter tuning was conducted via random search with 5-fold cross-validation, focusing on optimizing F1-score due to class imbalance. Model explainability was assessed using SHAP.
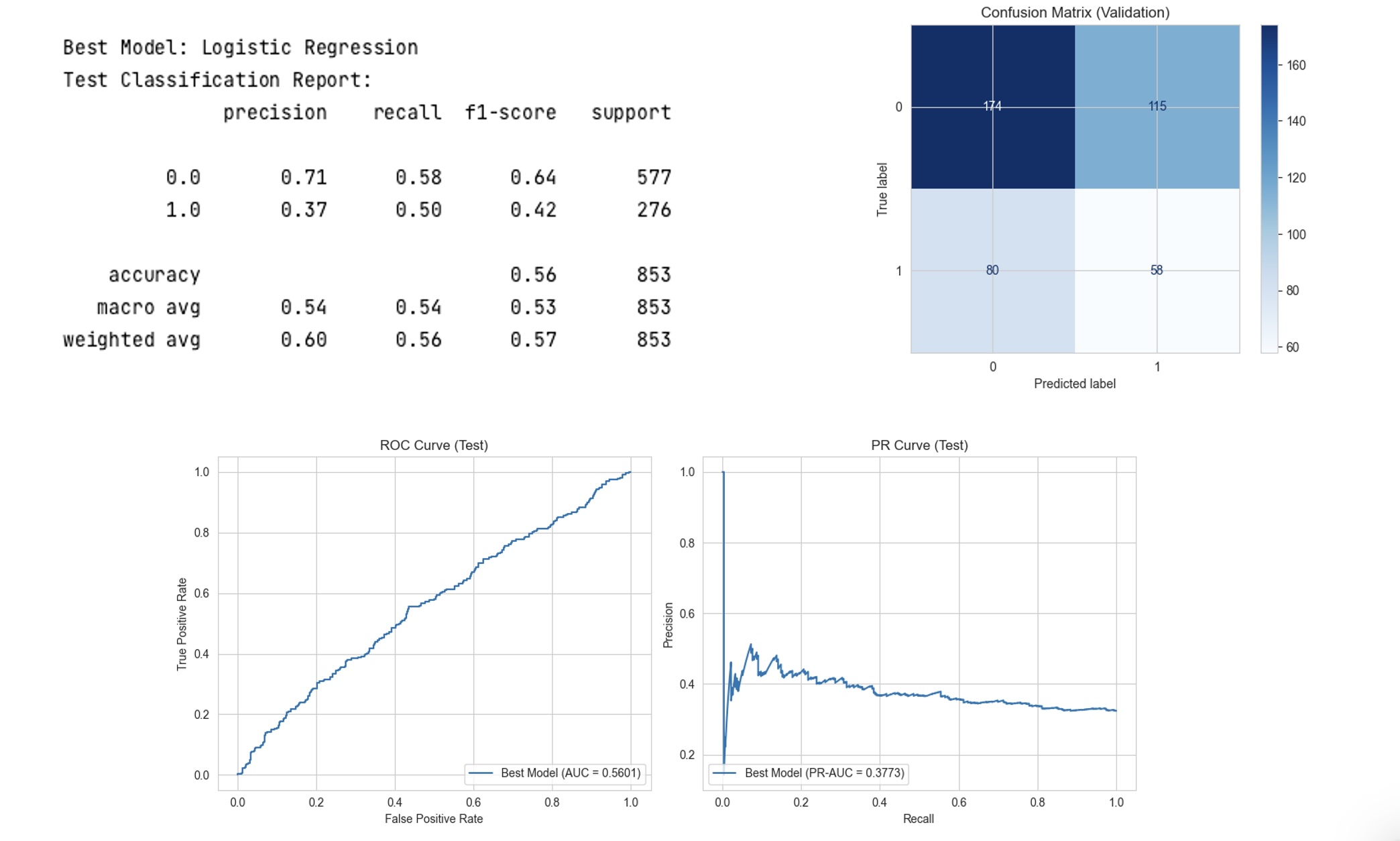
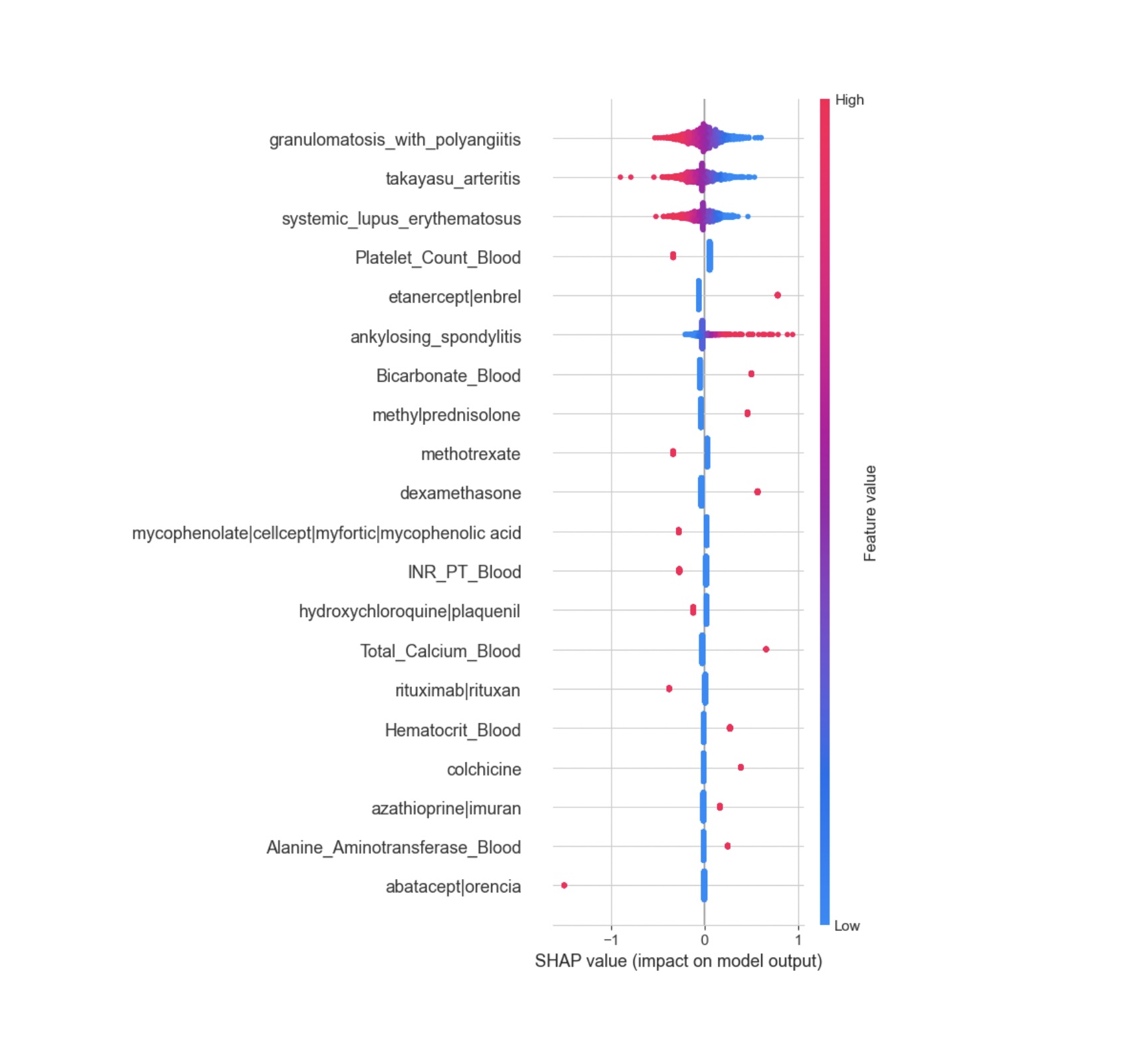
Results: The optimal Logistic Regression model, with L2 regularization, achieved superior performance with F1, recall, and precision scores of 0.52 on the validation set and 0.53 on the test set. The SHAP summary plot identified key features impacting model predictions, including specific rheumatic diagnoses and the presence of immunosuppressive medications. The model demonstrated a balanced approach in predicting infections, minimizing false negatives, which is critical in this patient population.
Conclusion: The machine learning model developed shows promise in predicting the risk of infection in hospitalized patients with rheumatic diseases. Key predictors included laboratory results, rheumatic diagnoses, and immunosuppressive medications. Despite limitations such as class imbalance and missing vital signs, the model serves as a proof of concept for using machine learning to assist clinicians in differentiating between infection and disease flare in this complex patient group. Future work will focus on improving model performance and incorporating patient-reported symptoms using natural language processing.
To cite this abstract in AMA style:
Felix M, Osmani L. Predicting Risk of Infection in Hospitalized Patients with Rheumatic Diseases from the MIMIC-IV Clinical Database: A Machine Learning Approach [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/predicting-risk-of-infection-in-hospitalized-patients-with-rheumatic-diseases-from-the-mimic-iv-clinical-database-a-machine-learning-approach/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/predicting-risk-of-infection-in-hospitalized-patients-with-rheumatic-diseases-from-the-mimic-iv-clinical-database-a-machine-learning-approach/