Session Information
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Systemic Lupus Erythematosus (SLE) is a heterogeneous autoimmune disease that requires more specific treatments to target patient subsets. To support precision medicine approaches, we report herein on a molecular profiling approach to stratify SLE patients, as well as the creation of a cohort of virtual patients to predict the efficacy of an anti-interferon (IFN)-α monoclonal antibody in Cutaneous Lupus Erythematosus (CLE) across patient heterogeneity. Systemic Lupus Erythematosus (SLE) is a heterogeneous autoimmune disease that requires more specific treatments to target patient subsets. To support precision medicine approaches, we report herein on a molecular profiling approach to stratify SLE patients, as well as the creation of a cohort of virtual patients to predict the efficacy of an anti-interferon (IFN)-α monoclonal antibody in Cutaneous Lupus Erythematosus (CLE) across patient heterogeneity.
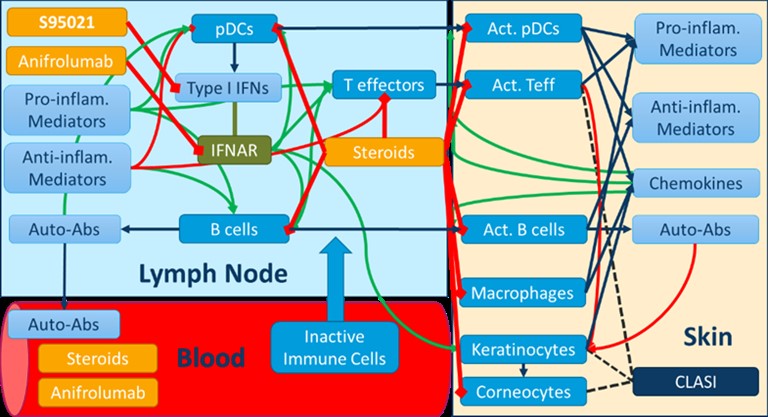
Methods: Multiomics profiling data from whole blood samples of 300 SLE patients and 330 matched healthy controls from the PRECISESADs cohort were used to stratify patients through hierarchical and k-means clustering. A quantitative systems pharmacology (QSP) mechanistic model recapitulating skin manifestations of CLE was developed using proprietary and public literature data, and qualified using the Rosa model qualification method [1]. This model was used to assess the efficacy of the pan-neutralizing anti-IFN-α S95021 monoclonal antibody .
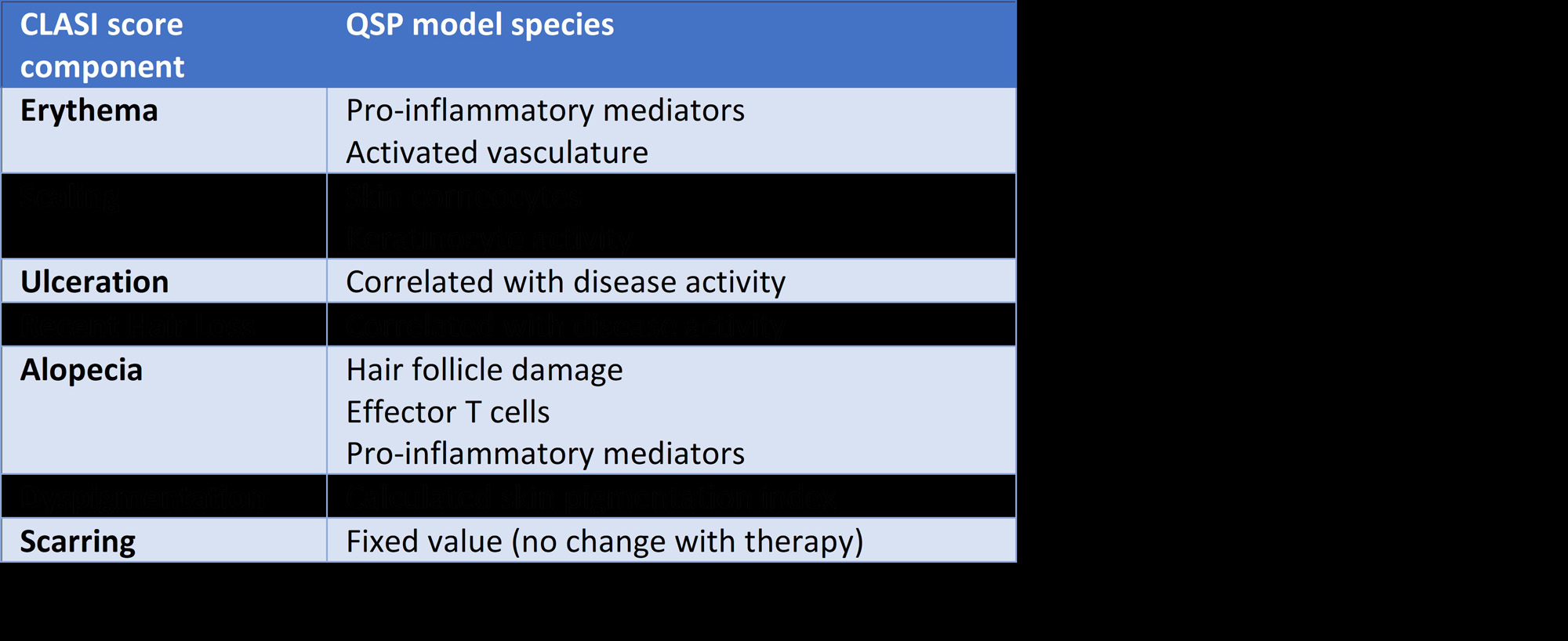
Results: The integrated analysis of multiomics profiling data led to the identification of four clusters of SLE patients. Gene enrichment analyses confirmed the upregulation of the type I IFN pathway in a majority of SLE patients, while further revealing distinct patterns of immune dysregulation involving B lymphocytes and autoantibodies, as well as T and polymorphonuclear cells. The QSP model of CLE relates various cellular and soluble proinflammatory mediators in the blood, lymph nodes and skin to subcomponents of the cutaneous severity score (CLASI). The time-course of CLASI score responses simulated in the model matched published clinical trial data with the anti-type I IFN receptor anifrolumab. Additionally, it predicted the efficacy of the S95021 antibody at various doses in CLE. By varying the contribution of the different cell types and mediators to skin damage and inflammation in the model, a cohort of ~700 virtual patients was generated, reflecting the heterogeneity observed across the various clusters of real patients from PRECISESADs. The analysis of predicted responders vs non responders using machine learning yielded biomarkers that could be used for the stratification of patients in subsequent clinical studies.
Conclusion: Combining molecular profiling of large cohorts of actual patients with QSP simulation encompassing hundreds of virtual patients is a powerful approach to document patient heterogeneity in a complex autoimmune disease. It also supports precision medicine strategies by enabling the creation of predictive models of drug response at an individual patient level. 1. Friedrich, C.M. et al, A model qualification method for mechanistic physiological QSP models. CPT Pharmacometrics Syst Pharmacol, 2016. 5: 43-53
To cite this abstract in AMA style:
Hurez V, Dasika K, Soret P, Myers R, Kudrycki K, Sheehan R, Friedrich1 C, Hubert S, Reed M, Desvaux E, Aussy A, Laigle L, Chadli L, Chassereau F, Fouliard S, Gauderat G, Moingeon P. Predicting in Virtual Patients the Efficacy of an Anti IFNa Mab in Cutaneous Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/predicting-in-virtual-patients-the-efficacy-of-an-anti-ifna-mab-in-cutaneous-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/predicting-in-virtual-patients-the-efficacy-of-an-anti-ifna-mab-in-cutaneous-lupus-erythematosus/