Session Information
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Deucravacitinib (DEUC) is a first-in-class, oral, selective, allosteric inhibitor of TYK2. TYK2 is required for signal transduction downstream of cytokines implicated in SLE pathophysiology, including type I and III interferons (IFN), IL-12 and IL-23. In a phase 2 trial (NCT03252587), DEUC was effective vs placebo (PBO) in patients with SLE receiving standard background therapy. Lupus is a disease with activation of innate and adaptive arms of the immune system, including pathways driven by IFN and B cell activity. Inhibition of TYK2 is expected to reduce the activity of SLE (ie, IFN-activation), however, the role of additional TYK2-regulated cytokines in SLE is not clear. We performed global transcriptomic analysis of patients treated with DEUC to further explore the mechanism of action (MOA) and effects of DEUC in SLE.
Methods: Blood samples (Paxgene RNA) were collected at baseline and multiple timepoints throughout the study following patient randomization (N=363). In a sub-study, samples (n=80) were collected 22-114 hours after the first dose. Global gene expression was assessed by RNA-sequencing and specific genes of interest were measured by endpoint PCR (Dxterity). Following quality control, 16,725 protein-coding genes were analyzed from 3,334 samples from patients with SLE. A set of demographically matched (sex and age) healthy control samples were collected to compare with study groups. Statistical analysis was conducted using Dream package in R. Differentially expressed genes from disease association and pharmacodynamic analyses were further characterized by pathway enrichment using standard databases.
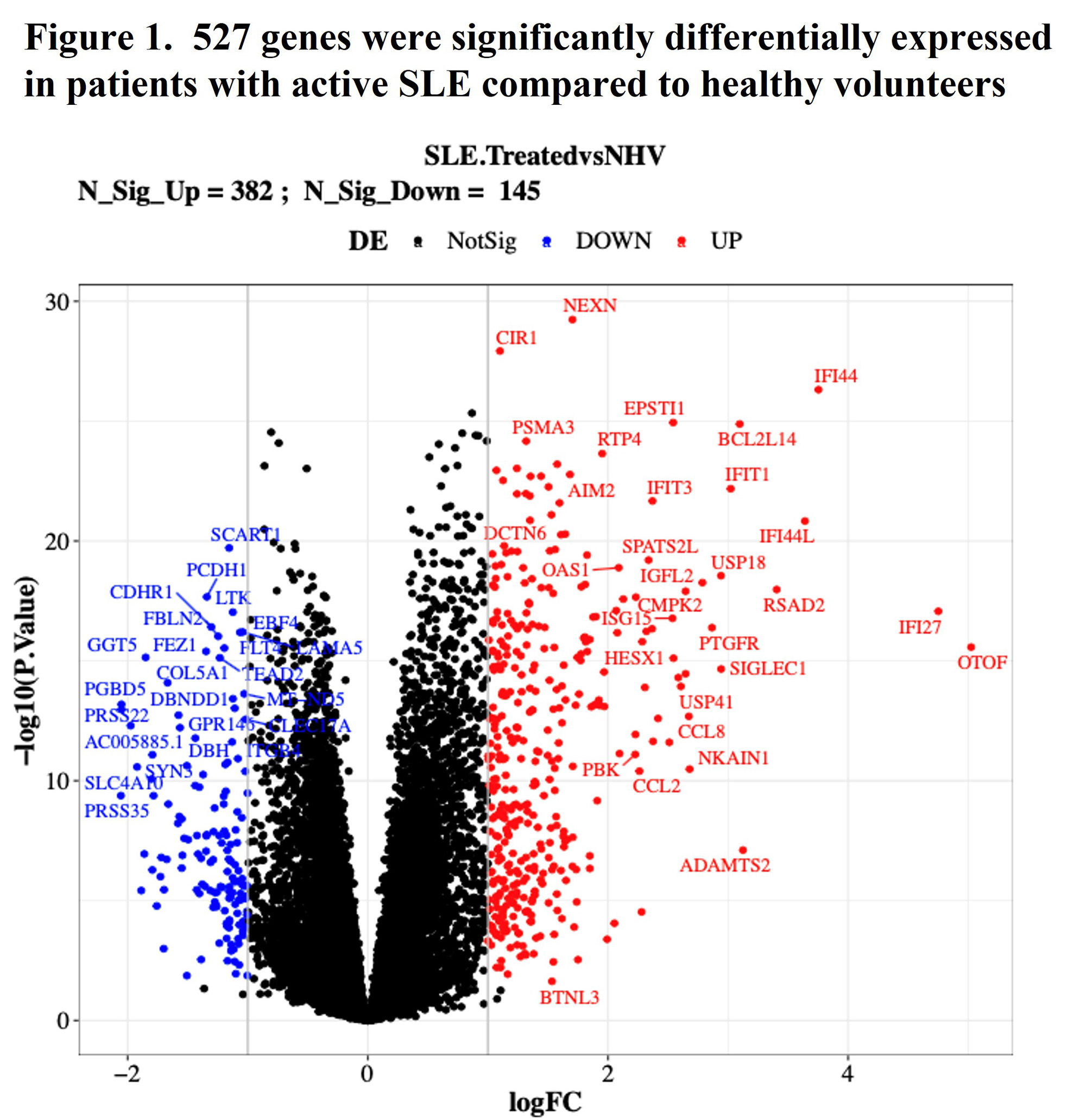
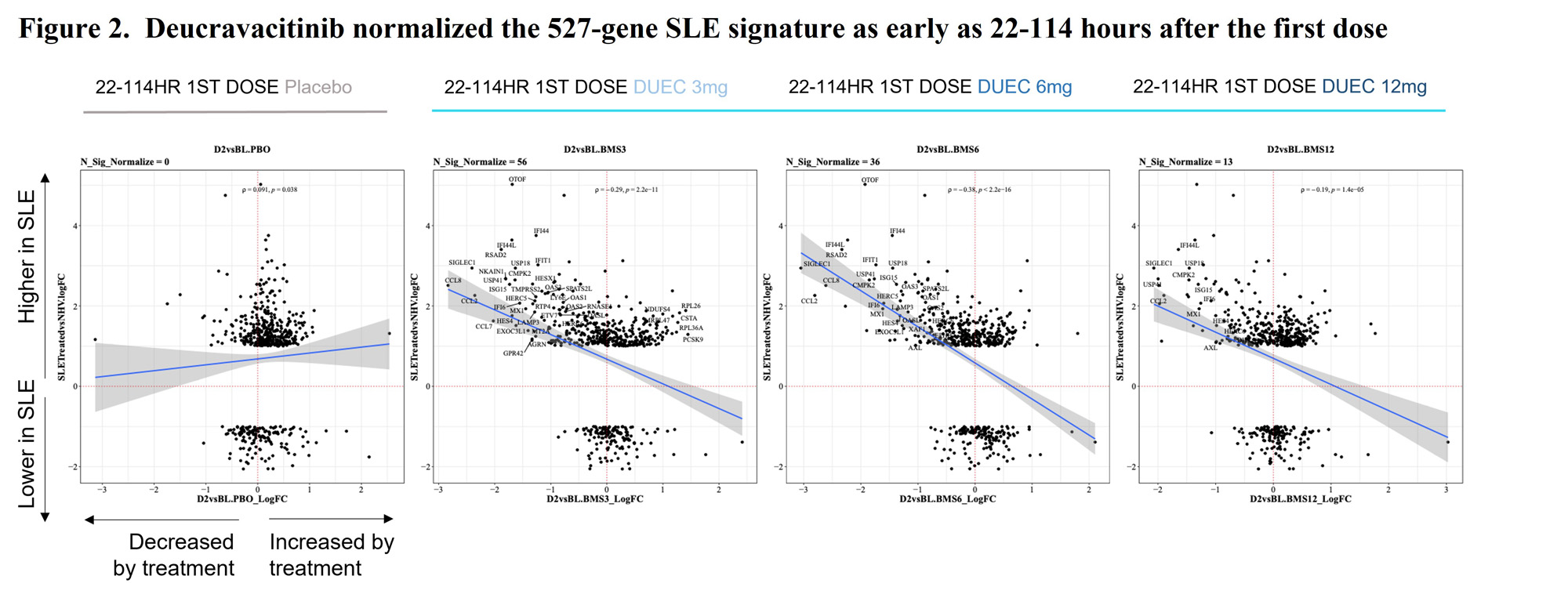
Results: 527 genes were significantly differentially expressed in subjects with active SLE compared to healthy volunteers with logFC >1 and adjusted P< 0.05 (Figure 1). The 527-gene signature was significantly enriched in 6 KEGG pathways, including “hsa05322-Systemic lupus erythematosus”. IFN-regulated genes were overexpressed as expected for patients with SLE. No gene expression changes were observed in PBO patients, however, treatment with DEUC significantly normalized the 527-gene set of differentially regulated genes. In the sub-study, gene expression was normalized in DEUC treated patients as early as 22-114 hours after the first dose (Figure 2). At the primary clinical endpoint of week 32, patients treated with DEUC exhibited significantly modulated gene expression of 461, 833 and 2529 genes, in the 3 mg BID, 6 mg BID, and 12 mg QD groups respectively.
Conclusion: Moderate to severe SLE is characterized by significant elevation of IFN regulated gene expression and B cell autoimmunity gene expression pathways. DEUC treatment significantly reduced IFN-regulated gene expression. These findings are consistent with the expected MOA, which inhibits signaling at the type I and type III IFN receptors. These results support the anticipated MOA and novel inhibition of B cell autoreactivity pathways, increasing evidence of this therapeutic strategy in the treatment of SLE. Additional analyses will focus on additional SLE-specific pathways by deucravacitinib and whether data will support a wider array of indications.
To cite this abstract in AMA style:
Wu C, Arriens C, Kahlenberg J, Hu Y, Hobar C, Coles A, Catlett I. Pharmacodynamic Changes in SLE Relevant Gene Expression Induced by Deucravacitinib in Patients Enrolled in the Phase 2 PAISLEY Trial [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/pharmacodynamic-changes-in-sle-relevant-gene-expression-induced-by-deucravacitinib-in-patients-enrolled-in-the-phase-2-paisley-trial/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/pharmacodynamic-changes-in-sle-relevant-gene-expression-induced-by-deucravacitinib-in-patients-enrolled-in-the-phase-2-paisley-trial/