Session Information
Session Type: ACR Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose:
As the cost of genome-wide genotyping plummets, and biobanking efforts integrating medical records and genetics are rapidly expanding, many patients will have genotyping available in medical records prior to patient visits. The question emerges: how informative this data is in a rheumatology clinic.
This study tests the potential for genetics to assign prior probabilities for six synovitis phenotypes; ACPA positive rheumatoid arthritis, ACPA negative rheumatoid arthritis, systemic lupus erythematosus, ankylosing spondylitis, psoriatic arthritis and gout.
Methods:
Risk allele information was obtained from Immunobase and GWAS publications. The genetic probability (GP) for each phenotype was calculated using Bayesian theorem with the summation of the risk alleles times the risk effects of each genetic variant as the likelihood and the sex adjusted disease prevalence as the prior. The GPs were normalized so that the total of the six probabilities for each individual was one. If GPs are accurate we expect that the percent of individuals that have a disease tracks with GP.
I Benchmarking performance by simulation
We simulated a population of one million people with random genotypes using minor allele frequencies from the 1000 genome project. We randomly assigned phenotypic status based on the known genetic risk alleles, sex, and disease prevalence. We then scored all individuals with disease for posterior probabilities.
II Validation
We selected clinical cases on their ultimate diagnosis from BostonÕs Partners BIOBANK, using a rule-based algorithm based on clinical notes, lab tests and medication prescriptions.
Results:
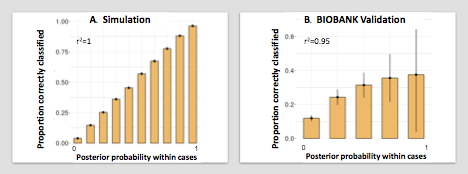
I In the simulation data set we observed that GPs were concordant with the phenotypic status, demonstrating that our model works in a theoretical dataset.
II We identified 297 out of the 15,000 patients in the BIOBANK that had one of the six diseases of interest. We used these individuals and assessed the performance of the GP.
We observed that there was a high positive correlation between the GP and the clinical case risk for the phenotypes (r2 = 0.95). A high GP correctly corresponded with the clinical phenotypes: in all cases where the probability > 0.9, the patient indeed had that phenotype. More importantly, the GP could discard phenotypes: GPs < 0.1 correctly corresponded with a clinical disease risk of < 10%, whereby for all individuals 1-4 phenotypes could be effectively ruled out. We note however that the GP (mean 0.24) somewhat overestimated the clinical risk (mean 0.16).
Conclusion:
In a cohort of synovitis patients, genetic information can facilitate decision making in early disease by ruling out and pointing towards the most likely phenotype. Seeing the importance of an early diagnosis in patients presenting with synovitis, genetics can be considered as part of a patientÕs medical history and as such it can inform us about the most likely diagnosis, without having to wait for more symptoms to arise.
To cite this abstract in AMA style:
Knevel R, Terao C, Cui J, Slowikowski K, Huizinga T, Karlson E, Raychaudhuri S. Optimizing Precision Medicine By Using Genetics to Assign Diagnostic Prior Probabilities to Patients with Synovitis [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 10). https://acrabstracts.org/abstract/optimizing-precision-medicine-by-using-genetics-to-assign-diagnostic-prior-probabilities-to-patients-with-synovitis/. Accessed .« Back to 2017 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/optimizing-precision-medicine-by-using-genetics-to-assign-diagnostic-prior-probabilities-to-patients-with-synovitis/